Indexed In
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- China National Knowledge Infrastructure (CNKI)
- Access to Global Online Research in Agriculture (AGORA)
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Publons
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
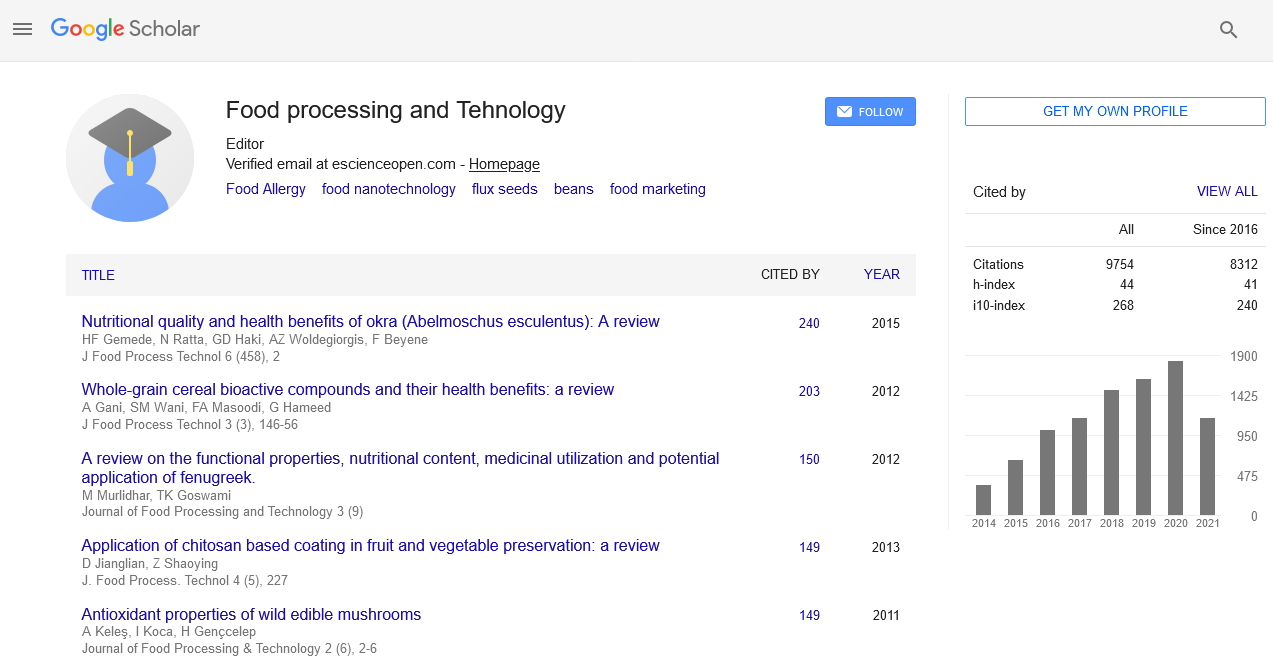
Marker-trait association analysis for grain calcium content in global collection of finger millet genotypes
Joint Conference on 8th World Congress on Agriculture and Horticulture and 16th Euro Global Summit on Food and Beverages
March 02-04, 2017 Amsterdam, Netherlands
Shambhavi Yadav, Anil Kumar and Ajay Thakur
Forest Research Institute, India
G.B. Pant University of Agriculture and Technology, India
Scientific Tracks Abstracts: J Food Process Technol
Abstract:
Statement of the Problem: Increased calcium content in cereals could result in biofortification for combating mineral deficiencies. Finger millet (Eleusine coracana) is one such crop with grains highly rich in calcium. Understanding the genetic basis of grain calcium content (GCC) is critical for improvement of finger millet. In present study, marker-trait association for GCC in finger millet was explored using association mapping. Methodology & Theoretical Orientation: A global collection of 238 genotypes was taken for present study and genotyped using 85 genic and non-genic simple sequences repeats (SSR) markers. Phenotyping for GCC was done using atomic absorption spectrophotometry. Association analysis of SSR allelic data with GCC was carried out using TASSEL 3.0.1 software. Results: The 85 SSRs yielded 160 scorable alleles, of which 62 were polymorphic with polymorphic information content ranging from 0.143 to 0.781 at an average of 0.359. GCC across 238 genotypes ranged from 0.07% to 0.45%. Association analysis revealed two genomic SSRs UGEP78 and UGEP60 to be significantly associated to GCC quantitative trait loci (QTL) at threshold (P) value‚?§0.001 and exhibiting phenotypic variance (R2) of 6.4 to 13.8% respectively. Genotypes GPHCPB45 and GPHCPB44 from North-West Himalayan region of India and exotic genotypes IE2957 and IE6537 were found to possess highest GCC. These may be effectively used as parents in genetic improvement programs. Conclusion & Significance: In present study, marker UGEP60 found linked to QTL for GCC belongs to linkage-group 1B of finger millet which shows co-linearity with rice chromosome 1 harboring a calcium content QTL and thus fine mapping around QTL on finger millet linkage-group 1B could lead to identification of potential genes affecting this trait. Moreover, identified markers could be used in map based cloning and marker assisted introgression. The present study therefore, provides a basis for genetic improvement of calcium content in finger millet.
Biography :
Shambhavi Yadav has her expertise in Molecular Plant Breeding. Her research work has been focused on “Use of molecular genetics and transcriptomics approaches in understanding calcium accumulation in finger millet”. She has done genetic diversity and population structure analyses in diverse finger millet germplasm and carried out linkage and association mapping to reveal quantitative trait loci for calcium content trait. Her work also include in silico mining of simple sequence repeats in finger millet developing-seed transcriptome thereby generating rich SSR resource in finger millet. Currently, she is working in the area of “Genetics and propagation of important forest trees and medicinal plants and also involved in in vitro conservation of forest genetic resources”.
Email: shambhaviyadav@gmail.com