Indexed In
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- China National Knowledge Infrastructure (CNKI)
- Access to Global Online Research in Agriculture (AGORA)
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Publons
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
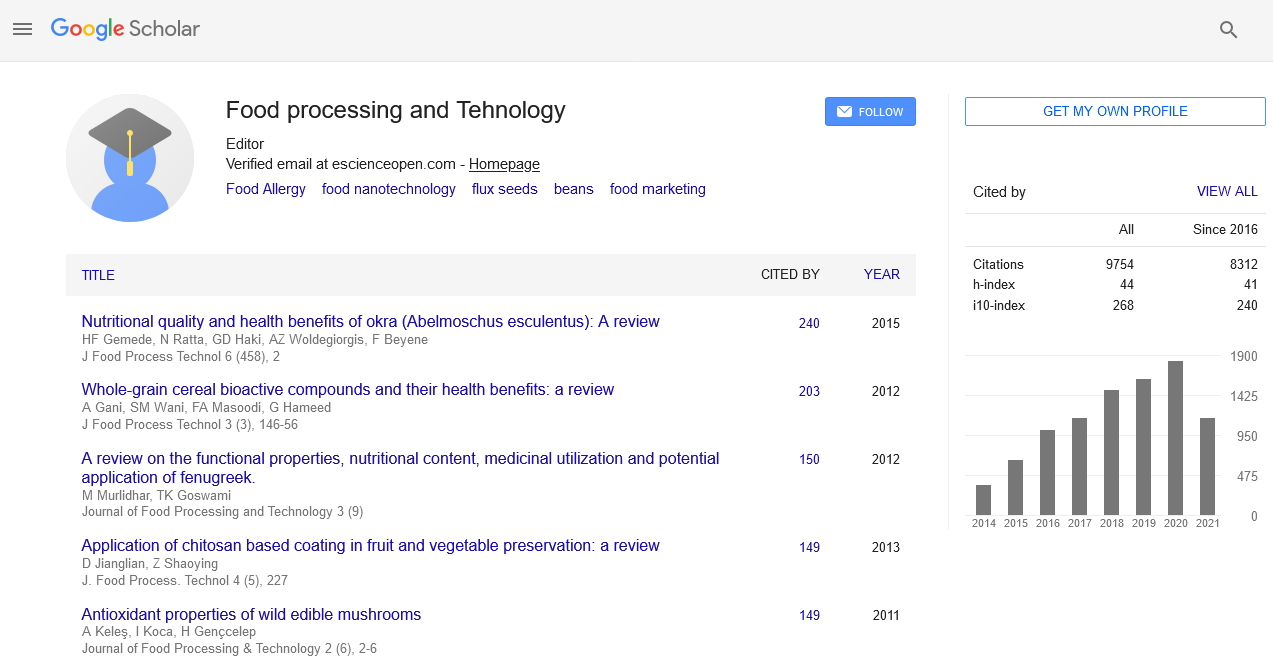
A universal molecular and bioinformatics pipeline for meat species identification using next generation sequencing data
9th Euro-Global Summit & Expo on Food & Beverages
July 11-13, 2016 Cologne, Germany
Nyaradzo Stella Chaora, KS Khanyile and FC Muchadeyi
Agricultural Research Council, South Africa
Posters & Accepted Abstracts: J Food Process Technol
Abstract:
Background: Processed meat products are usually targets of adulteration by fraudulent traders as a means of economic gain. This study demonstrates a universal and bioinformatics diagnostic pipeline using NGS data targeted for the 16S rRNA mitochondrial gene. Methods: A total of 161 processed meat products, namely, minced meat (n=55), biltong (n=28), burger patties (n=35) and sausages (n=43) were collected from retail outlets, butcheries and processing plants across South Africa. The MSR software managed to classify species up to the genus level. Results: The results revealed that 90% minced meat samples were predominantly Bos genus and 9% mince samples contained species from mixed sources like Bos, Sus and Ovis. About 92% of the biltong samples were predominantly Bos genus and 7% of the samples showed traces of Sus and Ovis genus, although they were labelled as ‚??beef biltong‚??. 80% of the burger patties were predominantly Bos genus, 20% were mixed samples carrying the Bos, Sus and Ovis genus, however, 5 of these samples were labelled as ‚??beef patties‚??. For the sausage samples 48% were predominantly Bos genus, 51% of the samples were mixed with traces of Bos, Sus and Ovis genus and of these 34% were labelled as ‚??beef sausages‚??. Conclusion: Overall, sausages had the highest occurrence of adulteration, constituting of species from more than one source in a sample.
Biography :
Email: ChaoraN@arc.agric.za