Indexed In
- Open J Gate
- Genamics JournalSeek
- Access to Global Online Research in Agriculture (AGORA)
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2023) Volume 12, Issue 2
Future of Stable Fermentation and Role of Next Generation Bio-sensors
Abu Feyisa1,2*, Mesfin Tafesse1, Musin Kelel1 and Gesesse Kebede12Department of Biology, College of Natural and Computational Sciences, Bule Hora University, P.O. Box, 144, Bule Hora, Ethiopia
Received: 09-Mar-2023, Manuscript No. FMT-23-20082; Editor assigned: 13-Mar-2023, Pre QC No. FMT-23-20082 (PQ); Reviewed: 27-Mar-2023, QC No. FMT-23-20082; Revised: 17-May-2023, Manuscript No. FMT-23-20082 (R); Published: 24-May-2023, DOI: 10.4172/2167-7972.23.12.167
Abstract
The objective of a bioprocess is to bring about particular chemical and/or physical changes in biochemical materials generated from the medium. Typically, a bioprocess will require fully functional live cells or enzymes as the biocatalyst. The possibility of unfavorable cell heterogeneity, which is defined by an increase in the frequency of low producing cells, is common during the scale up of bioprocesses. Low producing cells appear as a result of both genetic and non-genetic variation and will enrich as a result of their faster specific growth rate throughout the prolonged number of cell divisions involved in large scale bio production. The most recent methods for synthetic fermentation stabilization are presented, and it is argued that they should be used to create designs for cell factories that better meet industrial requirements. DNA sequencing information is used to guide strain design for standardized inoculum quality in bioreactors, with cell line screening and improvement by genome editing tools like CRISPR-Cas systems and genotype directed techniques. Furthermore, self-selecting phenotype directed techniques combine high output with cell proliferation, either by rerouting metabolic pathways or enriching for high performing cell variants using synthetic product bio sensing. Early evaluation of production stability in a new cell factory will direct design decisions that reduce variation. Production half-life from standardized serial passage stability screens and production load, measured as the production related percent wise growth rate reduction, serve as good initial measurements. Future cell factories will be far more scalable due to the incorporation of genetic designs that can maintain a high level of output over an extended period of time. Automated bioreactors are currently the most advanced bioreactors for longterm fermentation processes (automated stirred-tank and DASbox mini bioreactors: Parallel process development). Additionally, the use of next-generation biosensors for real-time particle monitoring, image processing, and imaging to monitor fermentation processes has made it possible for stable fermentation in this century.
Keywords
Genetic heterogeneity; Production load; Production stability; Phenotypic heterogeneity; Bioprocess
Introduction
Microbial fermentation has been used to successfully produce various compounds, including fuels, pharmaceuticals, medications, and other materials [1]. Even yet, there are a number of issues that industrial microbiological operations frequently face. There is a need for novel approaches to reduce the development costs associated with scaling up bioprocesses to high volumes in order to accelerate the commercialization of the increasing number of advanced fermentations [2]. Large scale bio-manufacturing is frequently constrained by the production phenotype's long term stability. Toxicities and metabolic burdens add to the production load (defined as percent wise reduction in specific growth rate associated with production). As cultures are extended to industrial numbers of cell generations (>40 generations), this production burden selectively enriches for naturally emerging low producing subpopulations while depleting high output subpopulations [3].
Physiochemical heterogeneity is another characteristic shown in large scale bio production as a result of insufficient mixing gradients that affect the substrate, oxygen, and pH regulators. Cellular heterogeneity, on the other hand, arises spontaneously and is enhanced by selection. The two types of cellular heterogeneity that can be identified are genetic and phenotypic (non-genetic) heterogeneity. According to Rugbjerg and Sommer, non-genetic heterogeneity is driven about by the regulation, expression, and random distribution of genes and proteins to new cells, whereas genetic heterogeneity occurs from a variety of gene and strain specific mutation types [3]. Even when starting with a pure master clone bank originating from a single cell, fermentations can have heterogeneities of low or non-producing cell variants [4].
The growth rate reduction driven on by metabolic burden (such as biochemical depletions in cofactors, redox, charged tRNA, and ATP) and metabolic inhibitors such intermediate and end product toxicity is known as the specific production load, which causes both hereditary and non-genetic variability. The overall burden of production can be calculated using the percentage wise decline in a particular growth rate brought on by output (production-associated fitness cost). Through the use of genomics technologies, it is possible to obtain a functional understanding of the elements that contribute to production load and potential remedies.
Deep understanding of the mutational mechanisms (rates, kinds, and targets) that even simultaneously can dominate production heterogeneity has lately been made possible by deep DNA sequencing advancements. This has made it possible for us to measure, evaluate, and control genetic heterogeneity [5]. The major techniques used to measure non-genetic production heterogeneity include biosensors, flow cytometry, and microfluidic cultivations [6].
Additionally, a group of scientists from Denmark founders the next generation biosensor to enable intelligent and reliable bioprocess (fermentation). They have been effective in monitoring the processes of fermentation by imaging and image processing using the ocelloscope device from bio sense solutions and through real-time particle monitoring using SOPAT [7].
This method should eventually reduce both sorts of variation in industrial cultures' synthetic biology. Addictions to manufactured products and autotrophies, which link development to the production of products, have been made possible by this transformation. With different degrees of effectiveness, product addictions can link growth to production utilizing product or pathway sensitive biosensors that regulate growth using either constitutive essential genes or conditional selection genes [8].
For instance, the population's productive lifetime increases when the initial production load is reduced [4]. The situation is complicated by the fact that there is an intrinsic positive link between production loads (burden and product toxicity) and production titers, rates, and yields. Therefore, strain variations that initially performed better may end up performing worse over the course of several cell generations compared to lower producing, less loaded strains, and increases in production load may be induced by declines in titers, rates, or yields [9].
In order to improve the design of scalable strains, it is crucial to monitor and manage both the production load and the rate of spontaneous heterogeneity development. The titers, rates, and yields measurements as well as long term durability forecasts may be dependent entirely on the production load and production half-life. To better predict the process scalability of new pathway designs in academia and industry, regular comparable stability screens to measure production half-life and regular measurements of production associated decrease in specific growth rate (fitness cost) to infer production loads and predict stability early in the design build test cycles of future cell factories are required.
Materials And Methods
Inoculum preparation and monitoringCell-line screening: The aim of cell line screening aims to find the most suitable native cell line that is currently available for the synthesis of a specific bioactive substance or enzyme. The initial cell lines screens either come from in house cell libraries or from commercial culture collections. In addition, specific databases, such as those detailing microbial metabolic pathways can be used to identify cell lines that are capable of specific biochemical conversions or the synthesis of particular compounds. Alternatively, entirely new cell lines can often be identified by sampling and enrichment from the natural environment, a procedure often termed bio-prospecting (Table 1).
Table 1: Advantages and disadvantages of expressions host cell.
| E. coli | Yeast | Insect | Mammal | |
|---|---|---|---|---|
| Biomass | High | High | Medium | Low |
| Growth (doubling time) | Fast (~20 min) | Medium (~120 min) | Slow (~120 min) | Slow (~24 h) |
| Cultivation | Easy | Easy | Easy | Elaborate |
| Coast | Low | Low | High | High |
| Protein folding | + | ++ | ++(+) | +++ |
| Popular strains/cell lines | E. coli BL21 | P. pastoris S. cerevisiae | Sf9, Sf21 | CHO, NSO, HEK293 |
Following the first screening phase, a few of the most promising cell lines are selected to proceed on to the cell-line enhancement phase of the development process. It is possible to precisely improve the performance of the native cell lines using a number of gene editing tools and molecular biology technologies. Since wild type cell lines often grow slowly and have a low yield of substances or enzymes on biomass, this augmentation stage is nearly always required.
Genome editing tools: Numerous genomes editing techniques, such as the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR-CAS system), Transcription Activator Like Effector-Based Nuclease (TALEN), and Zinc Finger Nuclease technology (ZFNs), have been developed in recent years to effectively modify multiple target genes in the genome [10]. The CRISPR-CAS system is presently the most straightforward, adaptable, and precise tool of genome modification. The initial discovery of the unique sequence of non-coding DNA known as clustered regularly interspaced short palindromic repeats, or CRISPR, in Escherichia coli was published in 1987 by Atsuo Nakata and his research team from Osaka university.
Biological function of CRISPR arrays was not known until 2005, when for the first time three studies suggested a role of it in adaptive immunity [11]. Then, in 2007, Barrangou, et al., provided evidence of adaptive immunity in bacteria by monitoring Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) loci in phage challenged cultures of Streptococcus thermophiles (one type of bacteria used to make yogurt and cheese).
In addition, reported that bacteria harboring a particular viral sequence as a CRISPR spacer were resistant to that virus, and that the CRISPR arrays were certified to provide protection against invading viruses when combined with Cas gene [12]. This refers to short palindromic sequence of DNA found within the genome of prokaryotic. Bacteria and archaea first developed CRISPR-Cas systems as a heritable form of adaptive immunity that protects prokaryotes from invasive heterologous DNA/RNA from viruses and other organisms and involved introducing a double-strand break at a particular target site that was complementary to crRNA or single-guide RNA (gRNA).
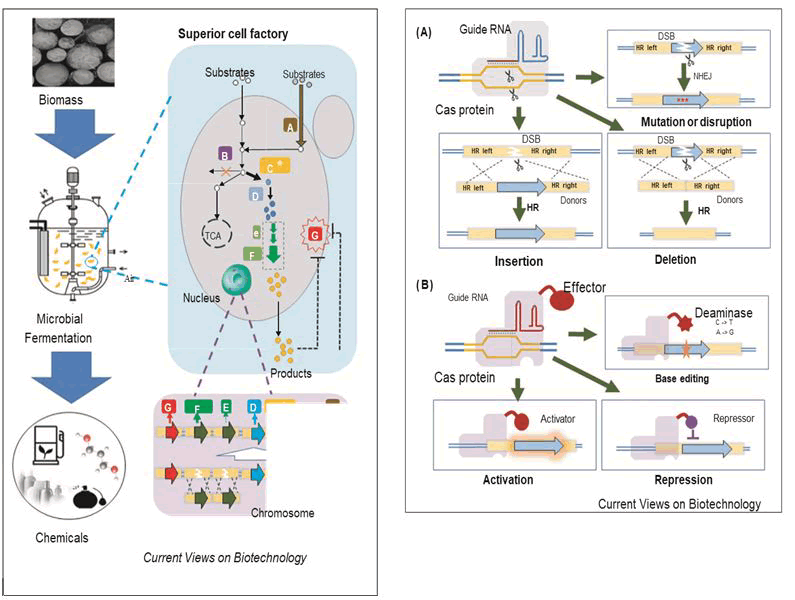
The application of CRISPR-Cas system was devised in 2012 when Jennifer Doudna a molecular biologist from university of California along with French microbiologist Emmanulle Charpentier, first proposed bacterial CRISPR-Cas system could be used as programmable toolkit of genome editing for human and other organisms and eventually receive the novel price in chemistry (CRISPR-Cas 9) in 2020 [13]. Liu, et al., constructed an endogenous subtype II-A CRISPR-Cas system based genome interference plasmids to exert high efficiency marker less gene deletion, gene integration, and point mutation in Pediococcus acidilactici [14]. Using this method, they found that the depletion of the native plasmids would increase cell growth, and the integration of an L-LDH gene into the genome would enhance cell growth and L-LA production (Figure 1).
Figure 1: Scheme of the CRISPR-Cas systems for genome editing and regulation. A) Basic applications of the CRISPR-Cas system for DSB-based genome editing, including gene deletion, mutation or disruption, insertion. In this system, a Cas nuclease (Cas 9) binds to a specific site in the genome guided by gRNA, and generates a DSB. Then desired genome editing could be achieved by HR or NHEJ. Donors are required when precise editing is conducted by HR; B) Advanced applications of the CRISPR-Cas system for non-DSB-based genome editing and regulation, including base editing, transcriptional activation (CRISPRa) and repression (CRISPRi). In this system, a dCas (dead Cas or nuclease-deficient Cas) is obtained by mutating a Cas nuclease, which can still recognize the binding sites but without the formation of DSB. Then, a customized function can be achieved by recruiting a desired effector (e.g. deaminase). (DSB) Double-Strand Break; (HR) Homology Repair; (NHEJ) Non-Homologous End Joining.
Predicting and quantifying production half‑lifeThe stability of new strains utilizing standard methodology will probably allow strain engineers to prioritize scalable early designs more effectively when compared to short term advantages that lack long term resilience. The direct production load can be predicted by comparing the relative specific growth rates of generating variants and low/non-producing variants, or the production half-life can be evaluated on serial passage stability screens [3].
Production half‑life estimation by serial passagingThe production half-life, which is estimated as the number of generations at which 50% of the initial production level is reached, can be determined via serial passing of cultures to replicate long term industrial cultivations. Seed trains are employed in industrial manufacturing to control cell doubling times and vaccinate massive fermenters. In the workplace, seed trains are typically passed to prevent lag phases following the stationary phase [15].
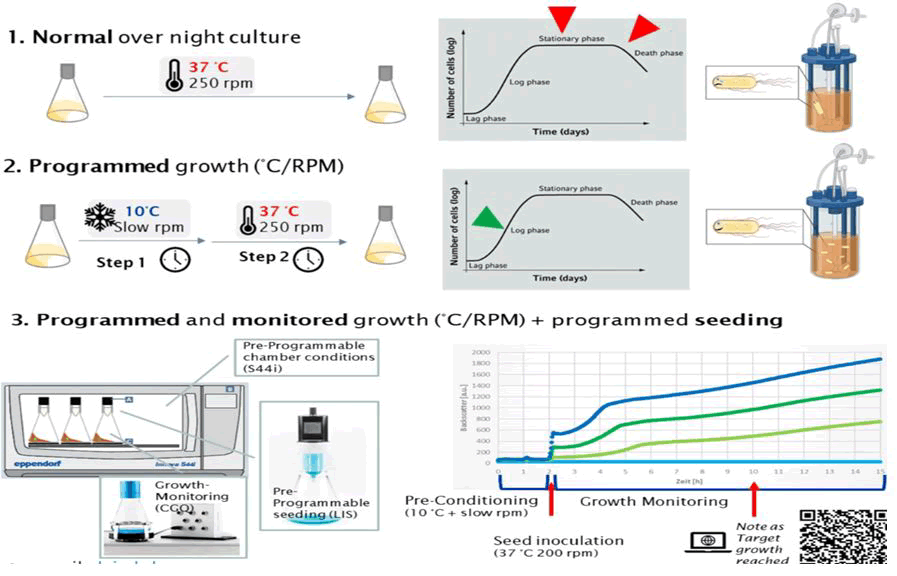
Intermittent stationary phases have direct effects on how future development will proceed, but in some cases, the stationary phase can also cause secondary mutational reaction [16]. Importantly, industrial practice can also comprise a number of processes, depending on the type of product, such as the use of inducers and feeding schedules that can significantly change the production burden (Figure 2). While it is important to avoid static moments while simulating scale up, cells are already under stress during the production process. Furthermore, the passage (seed) volume may have an impact on the simulation of long term cultivation if population bottlenecks are created by passing significantly fewer cells than the anticipated mutant generation rate [17].
Figure 2: Standardized inoculum quality for bioreactors and production half‑life estimation. Three hypothetical optimal scenarios are proposed in relation to, 1) Normal overnight culture (250 rpm at 37°C) cell are slowly grown, 2) Programmed growth (step 1 slow rpm and at so 10°C and step 2, 250 rpm at 37°C), 3) Programmed and monitored growth plus programmed seeding.
Factors affecting fermentation and methods of controlNon‑genetic and genetic heterogeneities: Phenotypic diversity among a population's individuals who have the same genome, as well as genetic heterogeneity mutations at two or more genetic loci that result in the same or similar phenotypes. Long term fermentations might be predicted to have a variety of heterogeneity kinds. Additionally, homogeneous alterations may potentially occur over time in cultures (by non-genetic inheritance of the proteome), which would have a greater impact on population dynamics than changes brought on by heterogeneity. However, cultural heterogeneity can potentially partially lower output levels depending on the available strategies for decreasing production load, as seen in early chemostat-based experiments [18,19].
Temporal variations in production performance may perhaps also be strictly homogeneous in addition to changes brought on by cell heterogeneities (developing non-producer cells) (uniform across the bulk population). A multi copy plasmid expressed form of S. cerevisiae was impacted by changes in protein composition during long term cultivation, such as insulin precursor synthesis [20].
Short term adaptation to the medium during propagation of the ethanol producing S. cerevisiae improves the quality of the propagated cells, improving overall ethanol productivity, suggesting a beneficial role for adaptation to the environment when producing a primary metabolite, such as by increasing cell viability [21]. As seen in populations of C. glutamicum that produce l-valine, non-genetic heterogeneity can reverse. L-valine concentrations were detected using a product biosensor connected to fluorescent protein [22].
Self‑selecting and phenotype directed strategiesRecent advances in synthetic biology have made it possible to stably stabilize high-performing fermentation populations by employing techniques that link a desired production phenotype to a certain growth rate at the single cell level. These techniques, which lead to the idea of "self-selecting fermentations," do not need to be aware of the precise escape routes because they aim to produce a certain product or significant intermediate along the metabolic process. Product addictions through genetically encoded product sensing biosensors or direct connection of production to necessary metabolism can both lead to production phenotype directed stability [23]. The product or intermediate is subsequently found by biosensors, who then use this signal to induce the production of genes essential for growth. Product addictions have been generated using conventional antibiotics or auxotrophy based selection genes as well as utilizing self-selecting, non-conditionally essential genes [24]. For several of the growth controlling genes utilized in studies, essentiality relies on the environment [4].
Self‑selecting fermentations by free mediumNon conditional (self-selecting) addictions regulate growth rates by constitutively essential genes rather than designing addictions using a possibly cross leaking metabolite or toxin [4,8]. Essential genes encoding for cell wall biosynthesis and antitoxins are used in self-selecting addiction systems (i.e. systems independent of media depletions/supplements). This establishes a link between the felt phenotype and a constitutive selection pressure in such addiction systems. Finding suitable constitutively necessary genes can be challenging. Regardless of the fact that they shouldn't have functional consequences on the metabolic production pathway, it is quite simple to screen for production cross talk. Constructing strains with constitutively addicted genes is challenging because constitutively essential genes prevent untangled selection. Self-selecting product addicted strains, on the other hand, promise longer stability and process adaptability by avoiding cross leakage and media conditioning. Both forms of product addiction systems add to the metabolic and transcriptional load, which may have a negative impact on the controlled cell factory strain's base performance. As a result, it will be crucial to evaluate and prevent any such stress caused by the system at the stage of development.
Bioreactor automationBecause advanced bioreactors improve productivity and remove unnecessary labor, time, and lab space for scientists who require large quantities of cells, microbes, or the products they express, they are effective instruments for addressing cultivation bottlenecks. The production of biopharmaceuticals, food and food additives, chemicals, and other goods is another area in which bioreactors are employed. They are also used in fundamental research and development. In bioreactors and fermenters, a wide variety of cell types and creatures can be grown, including cells (such as mammalian cell lines, insect cells, and stem cells), microorganisms (such as bacteria, yeasts, and fungi), plant cells, and algae [25].
Automated stirred tank: The bioreactor tank, which is also known as a vessel, is used for cultivation, and stirring instead of shaking is used to mix the culture. Stirred tank bioreactors are available in a variety of shapes and sizes, supporting cultures ranging from a few milliliters to millions of liters (usually glass, plastic or stainless steel). Stirred tank bioreactors always use the same fundamental parts and operate in the same way. Several components make up a stirred-tank bioreactor system (Figure 3). A container containing the medium used to culture cells, a vessel closing head plate, components, such as feed lines and sensors, inside the vessel or attached to the head plate to measure and modify the culturing conditions. A control system comprising external components used to adjust the culturing conditions (pumps) and control software [25].
Figure 3: Stirred tank bioreactor system including bioprocess control station, container, and software.
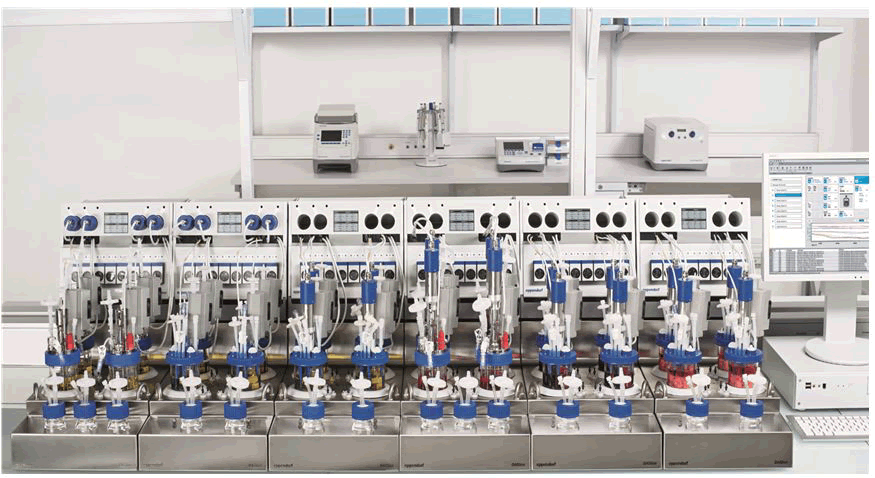
DASbox mini bioreactors: The DASbox is a revolutionary mini bioreactor device suitable for stem cell applications as well as microbial and cell growth. It is intended to operate as a 4-fold system with up to 24 parallel bioreactors. The DASbox is the ideal tool for sophisticated process research and design of experiment applications, with working capacities of 60 mL-250 mL. It is possible to carefully control every important process parameter. Easy handling with liquid free temperature regulation and exhaust condensation satisfies users [26]. Additionally, a single solution for all problems: Fermentation and cell culture, along with modest working volumes and parallel operation to speed up fermentation time to market, conserving lab space. The DASbox is suitable for microbiology and cell biology because of its comprehensive process control and precise monitoring of all key process parameters (Figure 4).
Figure 4: DASbox mini bioreactors system allows parallel operation of up to 24 bioreactors.
Results And Discussion
Future biosensor technology for sustainable fermentationsThe trend toward smart and stable fermentation in biotechnological processes is facilitated by latest innovations in sensors, process monitoring, and closed loop process control. This section's focus is to highlight two particular instances of clever solutions developed by the technical university of Denmark research team in close cooperation with commercial partners.
Image analysis and imaging based fermentation process monitoring: Measuring biomass and evaluating microbial growth are the two most obvious methods for evaluating the state of any fermentation. The microbial biomass, which is frequently the result itself, is the generating heart of any fermentation process [27]. However, the only practical choice for quantifying and measuring biomass is off line analysis. In the very early and late phases of fermentation, when there are, respectively, very low and very high biomass concentrations, on-line growth detection techniques perform less well. A contemporary accepted method for identifying growth is to evaluate Optical Density (OD). An increase in absorbance, which is typically tested at 600 nm, indicates that biomass development is progressing [28].
However, such measurements are inaccurate because of the constrained linear OD detection range of 0.1-1, which corresponds to a cell dry weight concentration range of approximately 0.1 g/l-1 g/l. The low reproducibility of spectrometers also has an effect on the results of such measurements. It is obvious that alternative methods that provide better detection of these crucial state metrics must be discovered in order to accomplish smart manufacturing. An optical window into the reactor has been made by recent developments in microscopic photography and image analysis, allowing for the automatic assessment of microbial growth as well as the observation of cellular processes.
Microscopy has grown into a potent tool that serves as the basis for advanced image-based surveillance and future control strategies. The imaging of living cells has greatly improved our understanding of cellular growth, functions, and responses to environmental changes, for example by revealing information about cell size, shape, position, and motility [29].
Phase contrast and confocal microscopy provides solutions to problems caused by various microscopic techniques, such as bright field and fluorescence microscopy that result in poor counting statistics, such as a small field of view or the visualization of objects that are transparent on the image, so called "phase objects." However, they are costly, demand specialized infrastructure, take a long time to acquire, and wrong settings might result in substantial artifacts on the photos. Bright field, digital, and time lapse microscopy are the foundation of the oCelloScope apparatus from bio sense solutions ApS in Farum, Denmark [30-33].
The device has a 200x magnification factor, making it possible to identify and segment objects between 0.5 m and 1 mm in size. As a result, it is appropriate for studying mammalian cells, yeast/fungi, bacteria, and crystals in (semi) transparent materials. Modern algorithms for automated image acquisition and thorough analysis are included in the software for operation and analysis. Using segmented images, objects are recognized by their actual shapes, and important characteristics like surface area, perimeter, and circularity are assigned. It has been demonstrated that the current open software platform offers significant benefits in a number of research domains, including microbiology and research. Moreover, the technology has found a solid application area in both monitoring of bacterial growth and growth inhibition, respectively, as well as for detection of changes in microbial morphology [34].
This method is especially inspiring since it can simultaneously detect morphological traits and assess microbial growth. For instance, it has been demonstrated that the cell viability (dead/alive, osmotically stressed, and the rate of the culture's growth) are all connected with information about cell size and cell size distribution in yeast cultivations [35]. Additionally, Marba-Ardebol, et al., found a correlation between cell size and the buildup of an internal product (fatty acids) in microalgae [36]. Therefore, it appears that image analysis is a potent technique that offers snapshots of the physiological condition through the evaluation of morphological aspects. Ultimately, image analysis can be used to assess the most crucial parameters such as microbial growth, substrate and product levels at a specific point in time, which forms the basis for novel image based control strategies for fermentation process operations.
Only off line sample analysis has been possible so far using the application, which is only compatible with microscopic slides and microtiter plates. On the other hand, real time, automated on line image data gathering and analysis are now possible thanks to particle tech ApS's recent creation of a flow through cell as an alternate sampling device. As a result, the first experiment of an online, image-based monitoring approach of a yeast fermentation process was set up using the produced prototype flow through cell by attaching the ocelloscope via a recirculation loop to the fermenter.
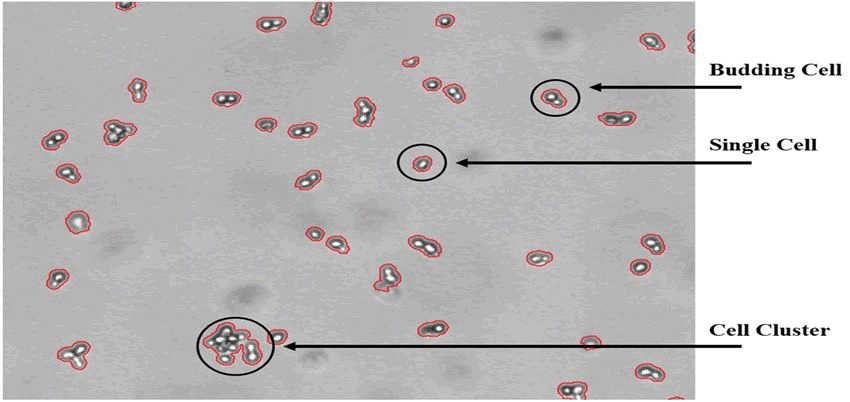
The objectives of the initial trial included the detection of growth and the assessment of morphological trends. The so-called bright spot feature, which takes advantage of the fact that yeast cells appear as a bright spot surrounded by a dark border on the images, was developed and included into the program to enable automated distinction of single cells, budding cells, and cell clusters. The bright spot approach enables the automatic differentiation between single cells, budding cells, and cell clusters by counting the number of bright spots associated with a cell object. Figure 5 shows an image of a yeast culture with various cell objects that was captured using the ocelloscope device. Figure 5 illustrates the bright spot feature's ability to operate consistently with an average failure rate of less than 5%. Based on a lab scale yeast fed-batch fermentation experiment (2l working volume, YPD medium, cultured at 30°C, 800 rpm, 1 vvm, controlled at pH 6), the validation results in Figure 4 indicate the addition of glucose (100 ml of a 400 g/l glucose solution) after 6 hours. Samples from the reactor were manually taken out and diluted to an OD of around 0.1 in parallel with the images taken with the ocelloscope device. In order to provide a suitable image quality with respect to the separation of cell objects, the yeast cell concentration associated with this OD value has previously been explored.
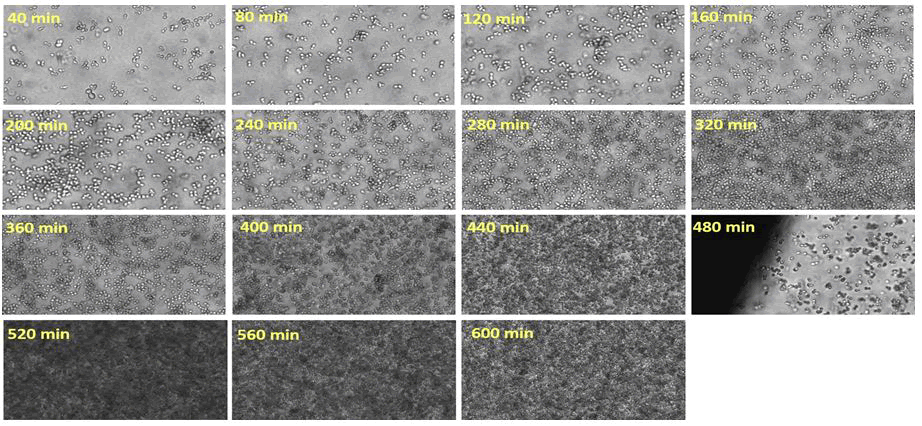
The findings of three photos after inoculation (0 min of fermentation time), after 240 min, and after 560 min of fermentation time are shown in Figure 5. Each image had an average of 1300 cell objects that were segmented. Following automated grouping of cell objects with 0, 1, 2, 3, 4, and 5 (or more) bright spots into corresponding groups, the groups were manually checked for false objects, and any false objects were manually removed from the corresponding group. Additionally, the bright spot feature has the ability to filter away image artifacts that come from background shading or out of focus cells, which leave objects with no bright spots (Figure 6).
Figure 5: Shows a yeast cell culture that has been developed in YPD media and has an OD value of about 0.1. A bright area with a darker border surrounds the yeast cells. The bright spot function keeps track of how many bright spots there are on each object. As seen in the figure, a single yeast cell is represented by a bright spot, a budding yeast cell is represented by two bright spots, and a cluster of yeast cells is represented by three or more bright spots.
Figure 6: 60% zoom into the images acquired on line, and the relative time point of image acquisition is indicated for each image. The big shading on the left corner on the image acquired at 480 min resulted from the border of an air bubble which was temporarily stuck inside the flow-through cell.
The prototype flow through cell application successfully illustrated the application of this technology for automated on-line growth detection. Every 10 minutes, photos were captured, and a sample of these images was acquired using the prototype flow-through cell. The upper detection was however restricted to an OD value of 4 (a cell dry weight of about 2 g/l) due to the chosen parameters (300 m depth of the prototype flow through cell). Online growth detection using the TA (Total Absorption) and BCA methods (background corrected absorption). Therefore, decreasing the liquid depth within the flow through cell lowers overall absorption and raises the maximum amount of biomass that may be detected [37].
For this reason, a new generation of flow through cells was created, allowing the system to handle a larger variety of cell concentrations during the fermentation process. These cells' depth may be modified automatically. The second development was a twin pump flow controller for automated dilution (Particetech ApS). This allows for the automatic diluting of samples from the fermenter, resulting in the proper cell concentration or, in turn, picture quality for segmentation. Together with the flow-through cell's height (depth) modification, this greatly facilitates the collection of online image data across a wider range of cell concentrations.
The ocelloscope device, a recent technical advancement, provides data on cell concentration as well as the dynamics of cell size, size distribution, and the dispersion of single cells, budding cells, and cell clusters. For instance, the latter could be connected to insulin manufacturing, which is crucial for yeast based insulin production systems. Additionally, researching how critical process events, such as stirring or aeration failure, which frequently pose operational challenges, affect cell morphology may have a substantial positive impact on integrated troubleshooting. These discoveries might inspire brand new, image based monitoring techniques for mass production.
Real time particle monitoring: Significant advancements in digital imaging, image analysis methods, and computing processing capacity have been made over the past ten years. This has made it possible to create new real time direct particle analysis techniques that use high resolution microscopic imaging to take pictures of particles suspended in liquid. One can quickly learn about the shape, size, and morphology-distributions of particles in a particle population by applying an image segmentation technique and then examining the recognized particles.
In the past 20 years, a variety of commercial particle monitoring options have been accessible. This includes non-invasive on-line flow-cell based sensors as well as in-line probe based sensors. With the latter technology, the imaging environment is more adjustable, which enhances image quality and has a significant impact on measurement accuracy. This includes superior illumination sources, less background noise due to the flow-thinness, cell's etc. Many commercial solutions have a detection limit that is lower, down to 0.5 m particles. This makes it possible to research a variety of particle in suspension based chemical and biological processes [24].
For instance, eukaryotic cells can be explored in fermentations, crystals in crystallizations, flocs in flocculation, and other processes. One may now monitor the dynamics of the particle processes with sampling times of less than a minute, and one can also use the particle analysis as a data source in process control techniques.
A number of various particle processes, including lab-scale crystallization, industrial scale crystallization, and a lab scale flocculation, have recently been examined by Gargalo, et al [7]. Here, they performed on line and at line sampling in turn, and the particle tech solution was used to examine the particles [24]. A peristaltic pump was used to transfer liquid samples from the reactor to an 800 m thick flow cell inside the particle tech microscopy equipment for the on-line observations. The liquid samples were imaged and then put back in the reactor [38-43].
Samples were directly transferred from the process tank to a microtiter plate for at line measurements, and the microtiter plate was subsequently put in the microscope unit for imaging. It was feasible to record the process dynamics as a set of time series data of particle characteristics and particle concentration by sampling every four to five minutes (Figure 7).
Figure 7: The process of image analysis, from imaging (left), through segmentation (middle) to analysis of individual particles (right). The particles are given random colors to indicate the particles detected during image segmentation.
Conclusion
Many innovations are probably going to increase long term bioprocess stability and production escape pathways, driven by current developments in synthetic biology, metabolic engineering, and next generation sequencing. More frequent assessments of the production burden (percent wise growth reduction caused by production) and the production half-life by serial passaging tests will be a crucial initial step. The burden and inhibitions underpinning the production load will be reduced through the application of particularly transformative methodologies, and synthetic biology tools will be used to control cellular production and growth. It is anticipated that both approaches focused on the mutant genotype and the production phenotype will help make altered production organisms more amenable to large culture volumes. Although it hasn't been demonstrated, self-selecting fermentations might theoretically also permit enriching for advantageous genetic variation. In order to increase output at the level of bulk populations, enrichment of advantageous genetic variants would probably take longer than the usual 100 generations of culture due to the comparably lower rates of spontaneous mutation than phenotypic change. As a result, this would seldom occur during industrial fed batch production, but the theory might be used to the creation of genetic variants that are more stable at the lab scale. It is becoming more and more feasible to create strains early on that will display long term stability as the areas of metabolic engineering and bio production expand with the emergence of numerous more sophisticated bio products.
Future work should enable more frequent assessment of the scalability of novel, promising strain designs and the extent to which homogenous long term adaptation in production organisms’ contrasts with widespread heterogeneity driven productivity decline. The exciting new advancements will offer continuous production a compelling alternative to the prevalent industrial fermentation processes already in use.
References
- Becker J, Lange A, Fabarius J, Wittmann C. Top value platform chemicals: Bio based production of organic acids. Curr Opin Biotechnol. 2015;36:168-175.
[Crossref] [Google Scholar] [PubMed]
- Wehrs M, Tanjore D, Eng T, Lievense J, Pray TR, Mukhopadhyay A. Engineering robust production microbes for large-scale cultivation. Trends Microbiol. 2019;27(6):524-537.
[Crossref] [Google Scholar ] [PubMed]
- Rugbjerg P, Sommer MO. Overcoming genetic heterogeneity in industrial fermentations. Nat Biotechnol. 2019;37(8):869-876.
[Crossref] [Google Scholar] [PubMed]
- Rugbjerg P, Myling-Petersen N, Porse A, Sarup-Lytzen K, Sommer MO. Diverse genetic error modes constrain large scale bio based production. Nat Commun. 2018;9(1):787.
[Crossref] [Google Scholar] [PubMed]
- Lin KY, Chen YC, Lin YF. LaMO3 perovskites (M=Co, Cu, Fe and Ni) as heterogeneous catalysts for activating peroxymonosulfate in water. Che Eng Sci. 2017;160:96-105.
- Xiao JP, He JF, Deng AP, Lin HL, Song T, Peng ZQ, et al. Characterizing a large outbreak of dengue fever in Guangdong Province, China. Infect Dis Poverty. 2016;5(1):1-8.
[Crossref] [Google Scholar] [PubMed]
- Gargalo CL, Udugama I, Pontius K, Lopez PC, Nielsen RF, Hasanzadeh A, Mansouri SS, et al. Towards smart biomanufacturing: A perspective on recent developments in industrial measurement and monitoring technologies for bio-based production processes. J Ind Microbiol Biotechnol. 2020;47(11):947-964.
[Crossref] [Google Scholar] [PubMed]
- Wang J, Zhang R, Zhang Y, Yang Y, Lin Y, Yan Y. Developing a pyruvate-driven metabolic scenario for growth coupled microbial production. Metab Eng. 2019;55:191-200.
[Crossref] [Google Scholar] [PubMed]
- Rugbjerg P, Olsson L. The future of self-selecting and stable fermentations. J Ind Microbiol Biotechnol. 2020;47(11):993-1004.
[Crossref] [Google Scholar] [PubMed]
- Lisby M, Rothstein R. Cell biology of mitotic recombination. Cold Spring Harb Perspect Biol. 2015;7(3):a016535.
[Crossref] [Google Scholar] [PubMed]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered Regularly Interspaced Short Palindrome Repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology (Reading). 2005;151(8):2551-2561.
[Crossref] [Google Scholar] [PubMed]
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. 2014;346(6213):1258096.
[Crossref] [Google Scholar] [PubMed]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. 2012;337(6096):816-821.
[Crossref] [Google Scholar] [PubMed]
- Liu L, Yang D, Zhang Z, Liu T, Hu G, He M, et al. High-efficiency genome editing based on endogenous CRISPR-Cas system enhances cell growth and lactic acid production in pediococcus acidilactici. Appl Environ Microbiol. 2021;87(20):e0094821.
[Crossref] [Google Scholar] [PubMed]
- Ikeda M. Amino acid production processes. Adv Biochem Eng Biotechnol. 2003;79:1-35.
[Crossref] [Google Scholar] [PubMed]
- Foster PL. Stress-induced mutagenesis in bacteria. Crit Rev Biochem Mol Biol. 2007;42(5):373-397.
[Crossref] [Google Scholar] [PubMed]
- Bull JJ, Barrick JE. Arresting evolution. Trends Genet. 2017;33(12):910-920.
[Crossref] [Google Scholar] [PubMed]
- Grzywacz D, Moore S. Production, formulation, and bioassay of Baculo viruses for pest control. Microb Control Insect Mite Pests 2017:109-124.
- Geng P, Leonard SP, Mishler DM, Barrick JE. Synthetic genome defenses against selfish DNA elements stabilize engineered bacteria against evolutionary failure. ACS Synth Biol. 2019;8(3):521-531.
[Crossref] [Google Scholar] [PubMed]
- Wright NR, Wulff T, Palmqvist EA, Jorgensen TR, Workman CT, Sonnenschein N, et al. Fluctuations in glucose availability prevent global proteome changes and physiological transition during prolonged chemostat cultivations of Saccharomyces cerevisiae. Biotechnol Bioeng. 2020;117(7):2074-2088.
[Crossref] [Google Scholar] [PubMed]
- van Dijk M, Erdei B, Galbe M, Nygard Y, Olsson L. Strain-dependent variance in short term adaptation effects of two xylose fermenting strains of Saccharomyces cerevisiae. Bioresour Technol. 2019;292:121922.
[Crossref] [Google Scholar] [PubMed]
- Mustafi N, Grunberger A, Mahr R, Helfrich S, Noh K, Blombach B, et al. Application of a genetically encoded biosensor for live cell imaging of L-valine production in pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum PloS one. 2014;9(1):e85731.
[Crossref] [Google Scholar] [PubMed]
- Wang X, Cabales A, Li Z, Zhang H. Biosensor-assisted high performing cell selection using an coli toxin/antitoxin system. Biochem Eng J. 2019;144:110-118.
- Liu L, Nakouzi E, Sushko ML, Schenter GK, Mundy CJ, Chun J, et al. Connecting energetics to dynamics in particle growth by oriented attachment using real time observations. Nat Commun. 2020;11(1):1045.
[Crossref] [Google Scholar] [PubMed]
- Meyer HP, Minas W, Schmidhalter D. Industrial‑scale fermentation. Ind Biotechnol: Prod proc. 2017:1-53.
- Silvia T, Vincent D, Jean-François H, Aurelie T, and Francoise D. DASbox Mini Bioreactor System, for microbial applications. Eppendorf Application Technologies. 2021.
- Zhang L, Zhang B, Zhu X, Chang H, Ou S, Wang H. Role of bioreactors in microbial biomass and energy conversion. Bioreactor Micr Biomass Energy Conv. 2018:39-78.
- Yang K, Zhang Y, Cai M, Guan R, Neng J, Pi X, et al. In vitro prebiotic activities of oligosaccharides from the by-products in Ganoderma lucidum spore polysaccharide extraction. RSC Adv. 2020;10(25):14794-14802.
[Crossref] [Google Scholar] [PubMed]
- Ettinger A, Wittmann T. Fluorescence live cell imaging. Methods Cell Biol. 2014;123:77-94.
[Crossref] [Google Scholar] [PubMed]
- Smith CL. Basic confocal microscopy. Curr Protoc Neurosci. 2011;56(1):2.
[Crossref] [Google Scholar] [PubMed]
- Telser A. Fundamentals of light microscopy and electronic imaging. Shock. 2002;17(5):442.
- Mondal PP, Diaspro A. Fundamentals of fluorescence microscopy: Exploring life with light. 2014. [Crossref]
[Google Scholar] [PubMed]
- Frigault MM, Lacoste J, Swift JL, Brown CM. Live cell microscopy tips and tools. J Cell Sci. 2009;122(6):753-767.
[Crossref] [Google Scholar] [PubMed]
- Fredborg M, Rosenvinge FS, Spillum E, Kroghsbo S, Wang M, Sondergaard TE. Rapid antimicrobial susceptibility testing of clinical isolates by digital time-lapse microscopy. Eur J Clin Microbiol Infect Dis. 2015;34(12):2385-2394.
[Crossref] [Google Scholar] [PubMed]
- Tibayrenc P, Preziosi-Belloy L, Roger JM, Ghommidh C. Assessing yeast viability from cell size measurements?. J Biotechnol. 2010;149(1-2):74-80.
[Crossref] [Google Scholar] [PubMed]
- Marba-Ardebol AM, Emmerich J, Neubauer P, Junne S. Single-cell-based monitoring of fatty acid accumulation in Crypthecodinium cohnii with three dimensional holographic and In situ microscopy. Process Biochem. 2017;52:223-232.
[Crossref] [Google Scholar] [PubMed]
- Campbell KL, Winters-Stone KM, Wiskemann J, May AM, Schwartz AL, Courneya KS, et al. Exercise guidelines for cancer survivors: Consensus statement from international multidisciplinary roundtable. Med Sci Sports Exerc. 2019;51(11):2375-2390.
[Crossref] [Google Scholar] [PubMed]
- He Y, Liu L, Zhu C, Guo S, Golani P, Koo B, et al. Amorphizing noble metal chalcogenide catalysts at the single layer limit towards hydrogen production. Nat Catal. 2022;5(3):212-221.
- Kjeldsen TS, Sommer MO, Olsen JE. Extended spectrum β-lactamase-producing Escherichia coli forms filaments as an initial response to cefotaxime treatment. BMC microbiology. 2015;15:63.
[Crossref] [Google Scholar] [PubMed]
- Shi S, Qi N, Nielsen J. Microbial production of chemicals driven by CRISPR-Cas systems. Curr Opin Biotechnol. 2022;73:34-42.
[Crossref] [Google Scholar] [PubMed]
- Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, et al. A communal catalogue reveals earth's multiscale microbial diversity. 2017;551(7681):457-463.
[Crossref] [Google Scholar] [PubMed]
- Van Breda AD, Theron LC. A critical review of South African child and youth resilience studies, 2009-2017. Child Youth Serv Rev. 2018;91:237-247.
- Wec AZ, Wrapp D, Herbert AS, Maurer DP, Haslwanter D, Sakharkar M, et al. Broad neutralization of SARS related viruses by human monoclonal antibodies. 2020;369(6504):731-736.
[Crossref] [Google Scholar] [PubMed]
Citation: Feyisa A, Tafesse M, Kelel M, Kebede G (2023) Future of Stable Fermentation and Role of Next Generation Bio-sensors. Ferment Technol. 12:167.
Copyright: © 2023 Feyisa A, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.