Indexed In
- Academic Journals Database
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- China National Knowledge Infrastructure (CNKI)
- Scimago
- Ulrich's Periodicals Directory
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- MIAR
- University Grants Commission
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2024) Volume 0, Issue 0
The Coviral Portal: Multi-Cohort Viral Loads and Antigen-Test Virtual Trials for COVID-19
Alexandra Morgan1, Elisa Contreras1, Michie Yasuda1, Sanjucta Dutta1, Donald Hamel3, Tarini Shankar1,4, Diane Balallo1,5, Stefan Riedel1,2, James E. Kirby1,2, Phyllis J Kanki3 and Ramy Arnaout1,2,6*2Department of Pathology, Harvard Medical School, Boston, MA 02215, USA
3Department of Epidemiology, Harvard T.H. Chan School of Public Health, Boston, MA 02215, USA
4Department of Pathology, Northeastern University, Boston, MA 02215, USA
5Department of Pathology, Boston College, Chestnut Hill, MA 02467, USA
6Department of Medicine, Beth Israel Deaconess Medical Center, Boston, MA 02215, USA
Received: 15-Jan-2024, Manuscript No. JVV-24-24621; Editor assigned: 18-Jan-2024, Pre QC No. JVV-24-24621 (PQ); Reviewed: 01-Feb-2024, QC No. JVV-24-24621; Revised: 08-Feb-2024, Manuscript No. JVV-24-24621 (R); Published: 15-Feb-2024, DOI: 10.35248/2157-7560.15.S25.001
Abstract
Objective: Regulatory approval of new over-the-counter tests for infectious agents such as SARS-CoV-2 has historically required that clinical trials include diverse groups of specific patient populations, making the approval process slow and expensive. Showing that populations do not differ in their viral loads the key factor determining test performance could expedite the evaluation of new tests.
Materials and methods: We annotated 46,726 RT-qPCR-positive SARS-CoV-2 viral loads with demographics and health status, evaluated the performance of two commercially available antigen tests over a wide range of viral loads, and created an open-access web portal allowing comparisons of viral-load distributions across patient groups and application of antigen-test performance characteristics.
Results: In several cases distributions were surprisingly similar where a difference was expected (e.g. smokers vs. non- smokers); in other cases there was a difference opposite from expectations (e.g. higher in patients who identified as White vs. Black). Predicted sensitivity and specificity of antigen tests for detecting contagiousness were similar across most groups.
Discussion: Rich clinical annotations reveal patient-subgroup-specific similarities and differences that are fertile ground for future research. Making viral loads freely and easily available for patient groups required significant attention to avoid potential loopholes that might risk patient privacy via identifiability. Two-parameter flexibility enables customized prediction of antigen-test results.
Conclusion: In silico analyses of large-scale, real-world clinical data repositories can serve as a timely evidence-based proxy for dedicated trials of antigen tests for specific populations. Free availability of richly annotated data facilitates large-scale hypothesis generation and testing.
Keywords
SARS-CoV-2; COVID-19; RT-qPCR; Viral load; Antigen test; Real-world data
Introduction
Diagnosing new pathogens requires developing diagnostic tests, which must be evaluated and approved by regulatory agencies before use for patient care. Such tests include Over-The-Counter (OTC) antigen tests, which have been widely used for at-home testing for COVID-19. To be approved, a new test must demonstrate a minimum level of clinical performance. Performance is typically measured as the test’s sensitivity (the proportion of true-positive samples that have a positive result) and its specificity (the proportion of true negatives that have a negative result). Clinical performance must be demonstrated in a defined patient population or group and clinical context, for example inpatients vs. outpatients. However, at the start of an outbreak, epidemic, or pandemic, there may not be enough information to know whether a test can be expected to perform differently by group. Therefore a new diagnostic test may be approved based on its performance in the general population, not specific groups.
Over time, evidence for clinical differences among specific groups may emerge. As this happens, it becomes reasonable to ask whether a test might perform differently across groups, with important implications for how and potentially even whether that test should be used for certain groups. Ideally, this question would be answered by conducting dedicated trials of the new diagnostic test in each group. Unfortunately, trials are expensive and slow. Also, the number of specific groups that may be of interest is large, since subgroups can be defined not only based on demographics (such as age, race, and gender), comorbidities (such as diabetes, heart disease, or immunosuppression), and care settings (inpatient vs. outpatient vs. emergency room) but also by combinations of these, an essential component of precision medicine. As a result, in practice it is prohibitively difficult to perform trials on specific groups for even a single diagnostic test, much less for the many tests that are likely to be developed in response to a large-scale outbreak, such as have happened during the COVID-19 pandemic. This is a problem for regulators, clinicians, and patients alike.
One solution is to apply a new test’s various performance characteristics to real-world data collected in the course of patient care. Such characteristics include results of existing trials as well as analytical (i.e. pre-clinical) operating parameters such as the Limit of Detection (LOD). The LOD is defined as the lowest concentration of virus that the test can detect in 95% of replicates. It is routinely determined by manufacturers and validated by clinical laboratories before a test is put to use clinically [1]. The relationship between concentration and detection is usually understood to follow an S-shaped curve [2]; fitting it requires at least one additional data point besides the LOD. The concentration of the virus may be measured as the viral load, most often defined as the number of copies of viral mRNA per milliliter of testing material (copies/mL).
“Real-world data” means the viral-load result of a reference diagnostic test that has already been approved for the general population. Because this data is from the general population, it will presumably include results on many specific patient groups. One can apply the performance of the new test as described above to the set or “distribution” of viral loads from a group to predict what proportion of patients in the group would have tested positive with the new test. This proportion is the sensitivity of the new test for that group. In this way, one can estimate clinical sensitivity without needing a dedicated trial on that group.
In this study, we apply this approach to COVID-19. We use the 46,726 positive SARS-CoV-2 RT-qPCR results our institution performed and use our electronic health record to annotate each result according to the patient’s demographics, comorbidities, and so on. Importantly, we convert each PCR result from a Ct value to a viral load using robust (100% code-coverage) and accurate publicly available software [1,3-5]. Ct values are less useful because they vary inversely with viral load and their scale differs by PCR testing platform.
We focus on sensitivity/specificity for contagiousness (i.e., infectivity). Contagiousness is of special interest given the public health focus on curtailing transmission [6,7]. Contagiousness can be estimated via a virus culture assay in which a positive patient sample is applied to susceptible cells and monitored for virus replication [8,9]. The lowest concentration of virus in a patient sample from which new virus can be recovered is the contagiousness threshold. Because cells in culture have no physical or distance barriers, mucociliary elevator, or protection via medications/immunity, we consider this threshold a conservative estimate. We previously demonstrated this threshold is approximately 50,000 copies/mL and has been fairly stable even as the SARS-CoV-2 virus has evolved [8,9].
Materials and Methods
See Supplementary Methods for complete details. Following institutional approval (BIDMC IRBs 2022P000328 and 2022P000288), specific patient groups were defined from the electronic health record based on prior work. Viral loads were obtained as described [3]. Select antigen tests and contagiousness were evaluated as described [8]. Presumed SARS CoV-2 variant was assigned using Covariants.org with confirmation of select strains by next-generation sequencing. Computational architecture and statistical tests are as described in the Supplementary Methods [10].
Results
A web portal for large-scale real-world SARS-CoV-2 viral load results for diverse patient groups
46,726 COVID-19 PCR results (as of April 14, 2023) representing approximately 39,180 unique individuals were converted to viral loads; annotated for patient demographics, comorbidities, presentation, treatment, and socioeconomic status; and made available for interactive investigation via a public web portal (Table 1). The portal [11,12] allows users to visualize the viral load distribution for any patient group, to compare distributions between groups, and to estimate, for each group, the sensitivity and specificity of a given OTC test for detecting contagious individuals. Users can define and compare complex subgroups by selecting multiple characteristics via checkboxes in the user interface (Figure S1).
| Patient's characteristics | COVID-19 PCR results |
|---|---|
| Sex | |
| Female | 25,884 |
| Male | 20,608 |
| Age | |
| <30 years old | 12,446 |
| 30-60 years old | 21,889 |
| >60 years old | 12,100 |
| Self-reported race or ethnicity | |
| Unknown/Other | 14,530 |
| White | 13,806 |
| Black | 8,299 |
| Hispanic | 7,540 |
| Asian/Pacific Islander | 2,472 |
| Setting | |
| Inpatient | 2,157 |
| Outpatient | 11,758 |
| Emergency room | 1,779 |
| Other institutions | 31,031 |
| Variant | |
| Early | 28,289 |
| Delta | 2,911 |
| Omicron | 11,264 |
| Vaccination status | |
| Vaccinated | 6,806 |
| Unvaccinated | 6,960 |
| Unknown | 32,732 |
| Outcome | |
| Died from COVID-19 | 398 |
| Died with COVID-19 as an incidental finding | 143 |
| Survived | 45,938 |
| Testing platform | |
| Abbott m2000 | 24,243 |
| Abbott Alinity | 20,593 |
| Abbott Alinity 4-plex | 1,889 |
| Total | |
| 46,726 |
Table 1: Summary of patient characteristics shown is counts for select high-level categories as of April 14, 2023. Counts may differ somewhat from the counts presented through the web portal as a result of jittering and as more data continues to be added through the portal. Note that counts broken down by characteristics do not add up to the total, because of the nulling out of some data to reduce re-identification risk (see Methods).
Overall viral load distributions
We found that viral loads varied over nearly ten orders of magnitude, from seven copies/mL (the lowest our system will report) to 1.5 billion copies/mL (99th percentile). This extraordinary range is consistent with observations from early in the pandemic (spring- summer of 2020) [1]. Those early observations suggested that viral loads were fairly uniformly distributed over the range. In contrast, the current dataset, which is ten times as large, demonstrates clear bimodality: viral loads were generally either very low, with a peak around the LOD of 100 copies/mL, or very high, with a peak~100 million copies/mL (Figure 1). This bimodality is apparent in retrospect e.g., in Figure 2A, of but required a large dataset to visualize clearly. Further research is needed to understand the reason(s) for these peaks [1].
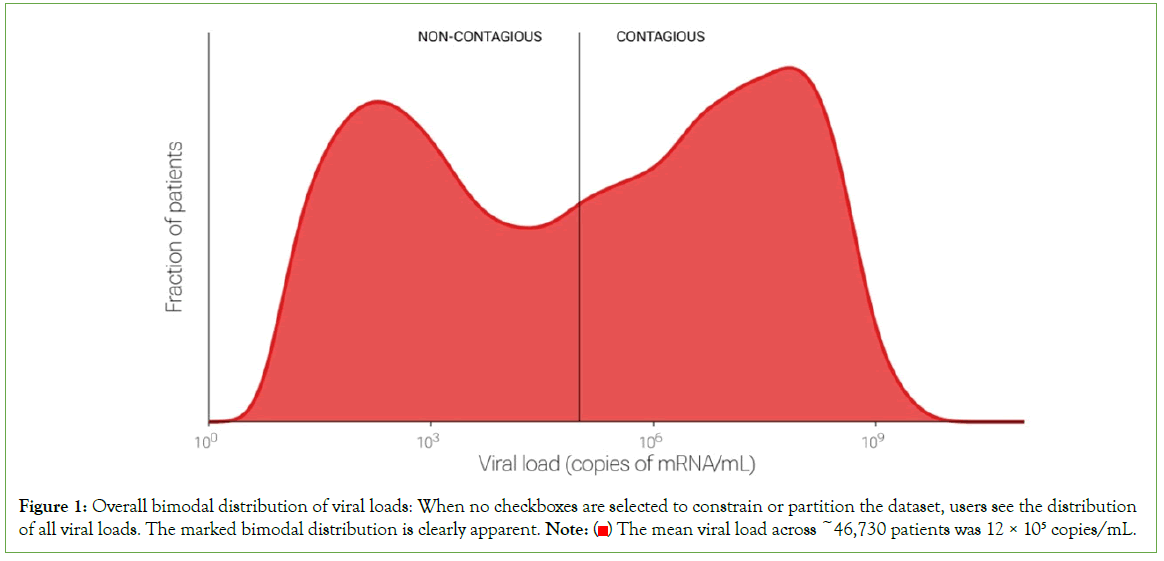
Figure 1: Overall bimodal distribution of viral loads: When no checkboxes are selected to constrain or partition the dataset, users see the distribution of all viral loads. The marked bimodal distribution is clearly apparent. Note:  The mean viral load across ~46,730 patients was 12 × 105 copies/mL.
The mean viral load across ~46,730 patients was 12 × 105 copies/mL.
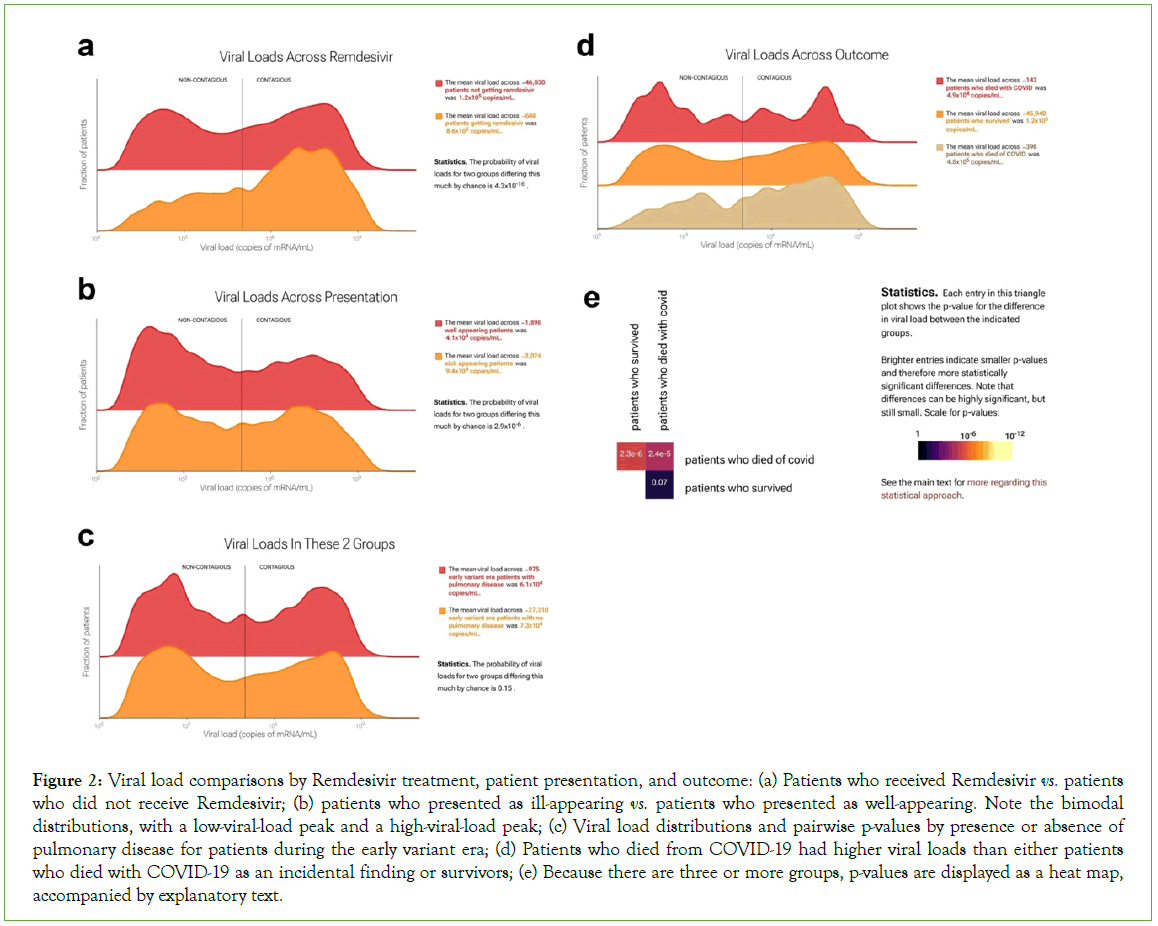
Figure 2: Viral load comparisons by Remdesivir treatment, patient presentation, and outcome: (a) Patients who received Remdesivir vs. patients who did not receive Remdesivir; (b) patients who presented as ill-appearing vs. patients who presented as well-appearing. Note the bimodal distributions, with a low-viral-load peak and a high-viral-load peak; (c) Viral load distributions and pairwise p-values by presence or absence of pulmonary disease for patients during the early variant era; (d) Patients who died from COVID-19 had higher viral loads than either patients who died with COVID-19 as an incidental finding or survivors; (e) Because there are three or more groups, p-values are displayed as a heat map, accompanied by explanatory text.
Viral load comparisons among patient groups-Remdesivir treatment and patient presentation:
The web portal allows statistical comparisons of thousands of specific patient groups. We describe several examples that are illustrative of the questions that can be asked and answered. Remdesivir (Gilead Sciences, Foster City, CA) is an intravenously administered RNA polymerase inhibitor initially approved by the FDA for treatment of SARS-CoV-2 in hospitalized adults and adolescents [13]. Of the 46,726 test results in our dataset, 688 were from patients who then received remdesivir. In practice, at our institution, remdesivir was used for sicker patients. We hypothesized that viral loads would be higher on average in patients who received remdesivir and in sicker patients (Figure 2). The portal supports this hypothesis: it shows viral loads were higher on average in both remdesivir-receiving and sicker-appearing patients, with means of 8.6 copies/mL × 105 copies/mL in patients who received remdesivir vs. 1.2 × 105 in those who did not (Figure 2A), and 9.4 × 104 in sick-appearing patients vs. 4.1 × 104 in well-appearing patients (Figure 2B). In both cases, the difference was due to greater fraction patients in the high-viral- load peak. In each case, the Kolmogorov-Smirnov [KS] p-value was 4.3 × 10-16, rejecting the null hypothesis of no difference. These are examples in which the portal facilitates confirmation of hypotheses regarding differences in viral load.
Unexpected findings-Pulmonary disease
Serious cases of COVID-19 are marked by life-threatening respiratory distress. This became less common with the emergence of the omicron strain and the increase in immunological exposure (via infection or vaccination). We hypothesized that patients with pulmonary disease would have higher viral loads than patients without it, especially for early viral variants, which had a stronger tropism for lung as opposed to the upper respiratory tract. However, the portal shows this hypothesis is not supported by the data (Figure 2C). Viral loads for the 975 patients with pulmonary disease tested during the early-variant era were statistically indistinguishable from those for the 27,308 patients with no pulmonary disease tested during the same time period. This is an example of unexpected findings that the portal can reveal.
Comparisons among multiple groups: survivorship and causes of death
The portal also allows users to compare>2 groups of patients at a time. For example, in quantifying mortality during the pandemic, one distinction of value has been between individuals who died from COVID-19 as the proximal cause of death vs. individuals who died with COVID-19 as an incidental finding. We used the portal to compare these two groups with survivors (Figure 2D).
We found that the 398 patients who died from COVID-19 had higher viral loads than either of the other two groups, and that viral loads were statistically indistinguishable between the approximately 46,000 survivors and the 143 patients who died with COVID-19 as an incidental finding (p=0.07). For ease of comparison, the web portal displays distributions in a ridgeline plot from lowest to highest mean, top to bottom. When there are three or more groups, p-values are displayed as a heatmap, accompanied by explanatory text. (Because KS p-values are symmetric, only the top half of the heatmap is shown.)
Complex patient subgroups: race and presumed variant
The ability to interrogate complex subgroups by checking multiple boxes in the web-portal interface allows more subtle investigations. For example, Black patients have experienced disproportionate morbidity and mortality during the pandemic [14]. However, the 13,806 patients who self-reported as white in our dataset on average had slightly higher viral loads than the 8,299 who self-reported as Black (KS p=3.3 × 10-13). That the viral loads in the white group were on average higher suggests that differences in outcome between these groups are not explained by differences in viral load (Figures 3A-3B), despite the clear relationship between viral load and survivorship described above (Figure 2D). Interestingly, the observed difference is more pronounced during the delta-variant wave (Figure 3C). During the delta wave (July to December 2021), viral loads were on average three times as high for White patients (8.0 copies/mL × 105 copies/mL, n=1,024) as Black patients (2.7 copies/mL × 105 copies/mL, n=665; KS test p=5.2 × 10-8) with a distinctly sharper high-viral-load peak in White patients. This difference was greater in patients over 30 years old and was almost entirely absent in patients under 30 (30-60 y.o: 398 Black patients and 719 White patients, p=2.5 × 10-5;<30 y.o.: 266 Black vs. 299 White patients, p=0.14). In contrast, viral load distributions for Black and White patients were more similar both early in the pandemic and during the omicron wave (p=4.0 × 10-5 for 4,393 Black and 7,042 White patients and p=0.02 for 2,295 Black and 4,341 White patients, respectively). This example illustrates the portal’s utility and (statistical) power for investigating complex subgroups and hypotheses.
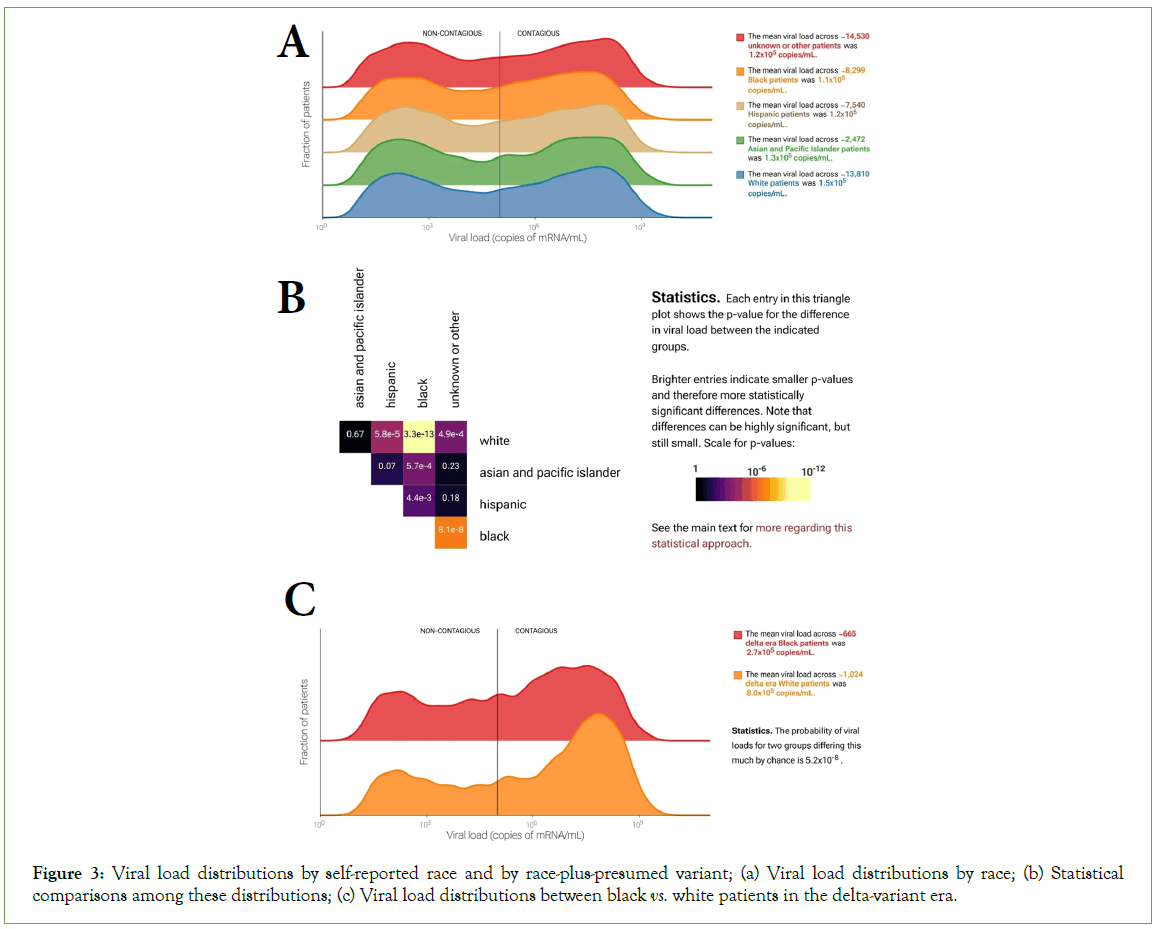
Figure 3: Viral load distributions by self-reported race and by race-plus-presumed variant; (a) Viral load distributions by race; (b) Statistical comparisons among these distributions; (c) Viral load distributions between black vs. white patients in the delta-variant era.
Antigen test performance
In the head-to-head comparison of PCR and antigen test results, 281 patients consented to participate. Of the 65 patients with positive PCR tests, 43 tested positive on the Binax antigen test and 40 tested positive on the CareStart antigen test. No invalid antigen tests (lacking the control line) were observed. Only one of the patients who tested negative by PCR tested positive on the antigen tests (both Binax and CareStart), confirming the high specificity of these tests. The proportion of positive antigen tests varied with viral load. At viral loads less than 103 copies/mL, both antigen tests were always negative; at viral loads greater than 107 copies/mL, both were always positive. However, there was an overlap of antigen- test-positive and antigen-test-negative results at intermediate viral loads (Figure 4A). k and v0 values (see Methods) were comparable between the two tests (k=1.184, v0=4.538 for Binax and k=1.142, v0=4.995 for CareStart). The resulting S-shaped curves were used to predict antigen test performance in the web portal.
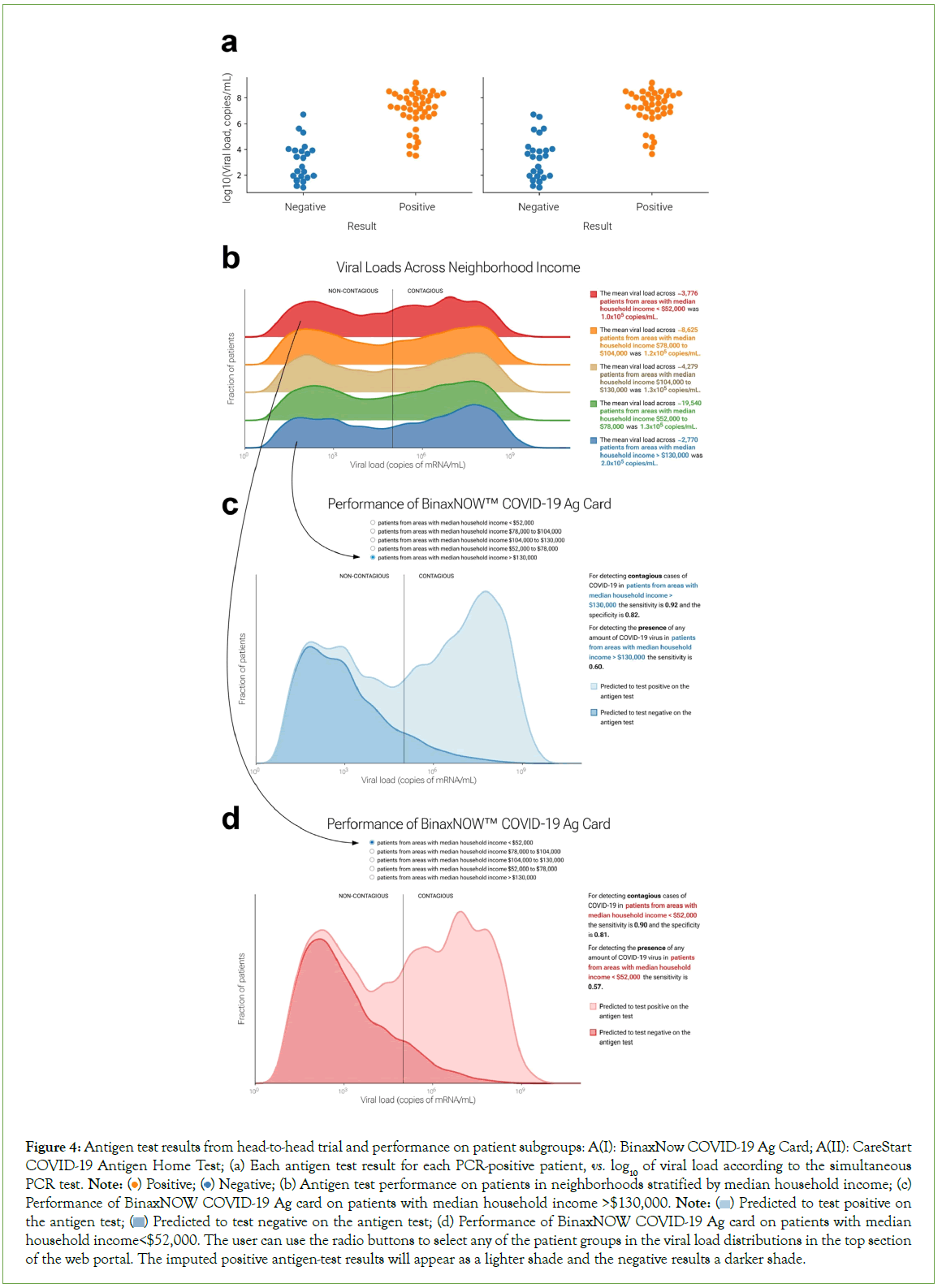
Figure 4: Antigen test results from head-to-head trial and performance on patient subgroups: A(I): BinaxNow COVID-19 Ag Card; A(II): CareStart COVID-19 Antigen Home Test; (a) Each antigen test result for each PCR-positive patient, vs. log10 of viral load according to the simultaneous PCR test. Note:  Negative; (b) Antigen test performance on patients in neighborhoods stratified by median household income; (c) Performance of BinaxNOW COVID-19 Ag card on patients with median household income >$130,000. Note:
Negative; (b) Antigen test performance on patients in neighborhoods stratified by median household income; (c) Performance of BinaxNOW COVID-19 Ag card on patients with median household income >$130,000. Note:  Predicted to test positive on the antigen test;
Predicted to test positive on the antigen test;  Predicted to test negative on the antigen test; (d) Performance of BinaxNOW COVID-19 Ag card on patients with median household income<$52,000. The user can use the radio buttons to select any of the patient groups in the viral load distributions in the top section
of the web portal. The imputed positive antigen-test results will appear as a lighter shade and the negative results a darker shade.
Predicted to test negative on the antigen test; (d) Performance of BinaxNOW COVID-19 Ag card on patients with median household income<$52,000. The user can use the radio buttons to select any of the patient groups in the viral load distributions in the top section
of the web portal. The imputed positive antigen-test results will appear as a lighter shade and the negative results a darker shade.
The OTC antigen tests that have been widely available on the market since 2021 are considerably less sensitive than RT-qPCR for detecting SARS-CoV-2 infection. However, because their LODs are generally above the contagiousness threshold, they are quite sensitive for detecting contagiousness [9].
Based on our clinical experience, we hypothesized that antigen tests would perform similarly on different patient groups and subgroups; this hypothesis was largely supported (Figures 4B-4D). The web portal also allows users to estimate sensitivity and specificity for the BinaxNow COVID-19 Ag Card and CareStart COVID-19 Antigen Home Test, based on the modelled performance curves, on any sufficiently large user-selected patient group (Figure 5). The two tests performed well: sensitivities for detecting contagiousness were roughly 0.85-0.90 across patient groups.
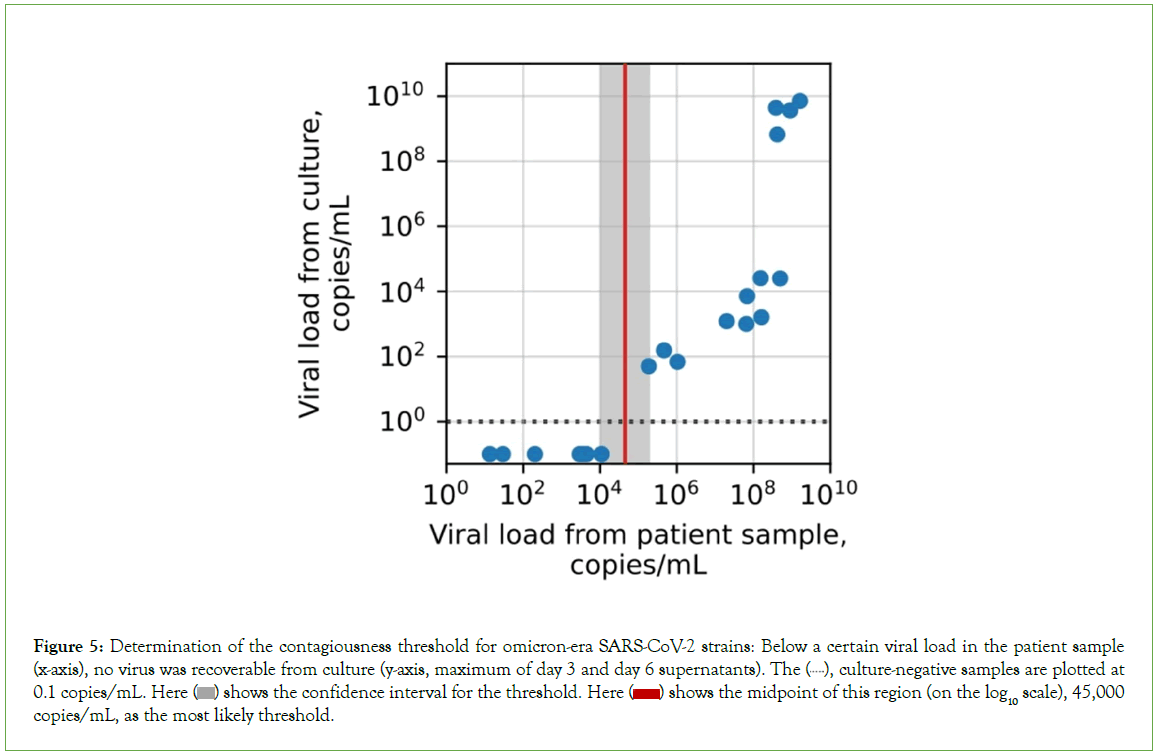
Figure 5: Determination of the contagiousness threshold for omicron-era SARS-CoV-2 strains: Below a certain viral load in the patient sample (x-axis), no virus was recoverable from culture (y-axis, maximum of day 3 and day 6 supernatants). The 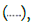 culture-negative samples are plotted at 0.1 copies/mL. Here
culture-negative samples are plotted at 0.1 copies/mL. Here 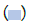 shows the confidence interval for the threshold. Here
shows the confidence interval for the threshold. Here  shows the midpoint of this region (on the log10 scale), 45,000 copies/mL, as the most likely threshold.
shows the midpoint of this region (on the log10 scale), 45,000 copies/mL, as the most likely threshold.
Contagiousness for omicron-era virus:
For early-pandemic and delta-wave strains of SARS-CoV-2, the threshold viral load for contagiousness has previously been found to be approximately 105 copies/mL [9]. For omicron variants, we found that the threshold is statistically indistinguishable from this, at 4.5 copies/mL × 104 copies/mL (confidence interval, 1.1 × 104 copies/ mL-1.9 × 105 copies/mL; p=0.23, Figure 4). The omicron threshold was based on 20 PCR-positive results from our head-to-head clinical trial of 277 total patients. We confirmed that the dominant strain circulating in the Massachusetts Bay area was omicron (BA5.2/ Clade 22B) via next-generation sequencing, with only rare single- nucleotide differences relative to those already described (Figures S2a-S2d). Because the strain mix in Massachusetts (Figure S2e), has been highly representative of the strain mix in the country as a whole throughout the pandemic (Figure S2f), these results support the generalizability of these findings from a particular geographic area to the entire population.
Discussion
The COVID-19 pandemic has proven a catalyst for accelerating medical advances, including the development of more efficient methods for developing and testing critical diagnostic assays [15-18]. It has also drawn attention to the value of reliable large public datasets [19-22]. Here we describe such a dataset, to our knowledge the first large dataset of SARS-CoV-2 viral loads in patients across the history of the pandemic through to the present. The portal revealed cases in which differences were expected as well as unexpected. The size of the dataset and extent of the annotation allow more comparisons than can reasonably be summarized in a single publication; the portal allows anyone clinicians, investigators, developers, regulators, and patients, alone or with the assistance of Artificial Intelligence and/or Machine Learning (AI/ML)-based tools to explore and conduct research, to test existing hypotheses, and to generate new research questions.
Access to the rich clinical annotations reveals similarities and differences in viral loads among patients by demographics, presentation, and comorbidity as well as by vaccination status, treatment, and socioeconomic status.
These similarities and differences are most properly understood not as cause-and-effect relationships but as practical descriptions, explanations for which are likely multi-factorial and fertile ground for future research. For example, living in a low-median-salary neighborhood should not be understood as causing higher viral loads, but people from such neighborhoods are more likely to have higher viral loads, however simple or complex the reason(s). Similarly, the bimodality of viral loads two peaks; one low, one high is also best interpreted through a practical lens as what is observed in clinical practice, with the full cause(s) deserving investigation.
The clinical utility of measuring and investigating viral load in SARS-CoV-2 has been amply demonstrated [23-26]. It is consistent with both the advantage of viral loads relative to Ct values [27,28] and their utility in earlier viral infections such as HIV and Hepatitis C (HCV). SARS-CoV-2 viral loads have proven useful in the development and characterization of COVID-19 diagnostics in multiple contexts, including testing on nasopharyngeal secretions, nasal secretions, and saliva [4,5]. Our results demonstrate their potential to regulators as a tool to streamline evaluation of new OTC tests. Specifically, a test’s performance as a function of viral load could be used to estimate that test’s clinical sensitivity for detecting contagiousness in any patient group, without having to conduct a dedicated clinical trial for that group. The alternative innumerable dedicated trials are unreasonable to expect, given the financial capabilities of developers and the bandwidth of their clinical testing partners. For this reason regulatory agencies such as the FDA have expressed interest in methods that use large-scale real-world data to streamline test evaluation.
Two assumptions implicit in this approach are worth mention. First, we model the success rate of antigen tests solely as a function of viral load, meaning we assume no other non-negligible factor varies systematically between patient groups. Second, we assume that the curve success rate as a function of viral load can be adequately predicted from a fairly small study. There are two possible sources of error in this curve: sampling error, which can be reduced by increasing the number of subjects sampled; and lack-of-fit error, the error inherent in trying to fit a function of the wrong form. The function used in the web portal’s calculations was chosen based on its long history in dose-response-type situations and comes with the (reasonable) assumptions that the probability increases smoothly and continuously with increasing viral load and approaches 0 with sufficiently low viral load and 1 with sufficiently high viral load. These constraints leave only two free parameters, which is desirable for statistical power and robustness to the noise inherent in any such study (e.g., sampling error and measurement error). These reasons rationalize its use.
A user of the portal who is accustomed to thinking of test quality solely in terms of LOD (Limit of Detection) might initially be surprised that the portal prefers two parameters to define antigen-test performance, not one. In effect, the LOD parameter sets the location of the S-shaped curve that relates viral load and performance, and the second parameter the 50% detection threshold sets the S-shaped curve’s steepness. Without the second parameter, one could fit a curve in which the sensitivity is 0% at all viral loads below the LoD and 95% at any higher viral load, which clearly would be quite different from the relationship observed in the head-to-head study. How different the true shape of the antigen test performance curve is from the logit function used here, and thus whether a different function would fit better, can be tested by larger head-to-head studies; however, in the midst of a public health emergency, the cost in time of sampling more subjects must be weighed against the value of complete certainty. Our fitting error was low.
Three limitations deserve mention the first is incompleteness of some of the data fields, for example presentation and vaccination status (Table 1). Presentation information was only sometimes available in structured form in our data repository; we did not attempt to extract data from notes to complement incomplete records. Vaccination status was likewise only sometimes available in a structured manner; integration with public-health records could potentially fill in missing records. Second, patient-level annotations are not yet available for download as part of the dataset. Making viral loads freely and easily available for patient groups required significant attention to avoid potential loopholes that might risk patient privacy via identifiability. Our methods included suppressing data transmission for groups small enough to present potential “journalist risk,” jittering counts to prevent deduction of the sizes of suppressed groups, and rounding viral loads to two log- scale decimal places [29,30].
Further work is necessary to make patient-level annotations available. Third, the size of the dataset, while large, is still insufficient to draw statistically strong conclusions for the smallest groups (e.g. Native Americans, cystic fibrosis patients, or puerperal mothers). One solution is to add data from other institutions that performed substantial COVID-19 testing; another is to supplement existing large datasets, for example the 50-million-person CVD-COVID- UK initiative, with viral loads. The free availability of methods to convert from Ct values to viral loads facilitates such advances [3].
Clinical care benefits from personalization of diagnostic testing: the right test for the right patient, where the importance of patient heterogeneity is increasingly accepted. Our portal demonstrates that large-scale real-world data can assist this effort by helping personalize test sensitivities and specificities without the need for dedicated clinical trials. This approach is generalizable beyond COVID-19. Laboratory testing is an exceptionally rich source of real-world medical information. It is the highest volume medical activity, with billions of tests performed annually worldwide. It is also the most cost-effective, costing just pennies on the healthcare dollar. It is integral to decision-making across medicine, for patients at every level of acuity, from screening to emergencies. Its results are almost always numerical or categorical, making it especially amenable to modern approaches like machine learning. And computational re-analysis is substantially less expensive than de novo trials. The present work supports the view that meaningful value can come economically from repurposing of the vast stores of real-world laboratory results for public use.
Data availability
Data for the plots in (Figures 1-4) our papers (except figure 4a) are reproducible. Deidentified data for figures 4a and figure 5 available upon request. Aggregate group-level data is publicly available via API from our web server. Deidentified patient-level data will be made available by arrangement, subject to IRB approval. Please contact the corresponding author to apply.
Conclusion
In conclusion, harnessing in silico analyses on vast clinical data repositories provides a practical and evidence-backed substitute for dedicated trials of antigen tests targeted to specific populations. The open accessibility of meticulously annotated data not only supports extensive hypothesis formulation but also expedites large- scale testing initiatives, offering a valuable resource for advancing diagnostic methodologies in a timely and efficient manner.
The COVID-19 pandemic has not only prompted significant medical advancements but has also underscored the importance of extensive public datasets. The presented dataset, spanning the entire pandemic, offers a comprehensive exploration of SARS- CoV-2 viral loads, revealing nuanced patterns influenced by demographics, vaccination status, and socioeconomic factors. While cautioning against oversimplification of observed correlations, the study highlights the clinical utility of viral load measurements in refining COVID-19 diagnostics and regulatory evaluations. The portal’s unique approach, leveraging large-scale real-world data, demonstrates the potential for personalized diagnostic testing, extending beyond COVID-19. The study advocates for the economical repurposing of abundant laboratory data to enhance public health.
Author Contributions
Conceptualization: JK and RA; Software: AM; Investigation: DH, EC, SD, and MY; Resources: SR; Data Curation: AM; Writing original draft: RA and AM; Writing, review and editing: PK and JK; Visualization: TS, DB, AM, and RA; Supervision: RA; Funding acquisition: PK, SR, and RA.
Acknowledgment
The authors acknowledge Elliot Hill for assistance on plugging ct2vl into the data processing workflow; to Timothy Graham, Gail Piatkowski, Baevin Feeser, and Griffin Weber for their advice and guidance on extracting data from EMR databases. This work was supported by the Reagan-Udall Foundation for the FDA (RA); and a Novel Therapeutics Delivery Grant from the Massachusetts Life Sciences Center (JEK). Abbott (Scarborough, ME) provided the Binax Now antigen tests, and Ginkgo Bioworks (Boston, MA) provided the CareStart antigen tests used in this study.
References
- Arnaout R, Lee RA, Lee GR. The limit of detection matters: The case for benchmarking Severe Acute Respiratory Syndrome Coronavirus 2 testing. Clin Infect Dis. 2021;73(9):3042-3046.
- Wild D, Editor. The immunoassay handbook: Theory and applications of ligand binding, ELISA, and related techniques. 4th ed. Oxford ; Waltham, MA: Elsevier 2013.
- Hill ED, Yilmaz F, Callahan C. ct2vl : Converting Ct values to viral loads for SARS-CoV-2 RT-qPCR test results. Microbiol. 2021:2022-2106.
- Callahan C, Rose AL, Rye LG. Nasal swab performance by collection timing, procedure, and method of transport for patients with SARS-CoV-2. J Clin Microbiol. 2021;59(9):10-128.
[Crossref] [Google Scholar] [PubMed]
- Callahan C, Ditelberg S, Dutta S. Saliva is comparable to nasopharyngeal swabs for molecular detection of SARS-CoV-2. Microbiol Spectr. 2021;9:e0016221.
[Crossref] [Google Scholar] [PubMed]
- Pilarowski G, Lebel P, Sunshine S. Performance characteristics of a rapid Severe Acute Respiratory Syndrome Coronavirus 2 antigen detection assay at a public plaza testing site in San Francisco. J Infect Dis. 2021;223(7):1139-1144.
[Crossref] [Google Scholar] [PubMed]
- Mina MJ, Parker R, Larremore DB. Rethinking COVID-19 test sensitivity-A strategy for containment. N Engl J Med. 2020;383(22):e120.
[Crossref] [Google Scholar] [PubMed]
- Stanley S, Hamel DJ, Wolf ID. Limit of detection for rapid antigen testing of the SARS-CoV-2 Omicron and Delta variants of concern using live-virus culture. J Clin Microbiol. 2022;60(5):e00140-22.
[Crossref] [Google Scholar] [PubMed]
- Kirby JE, Riedel S, Dutta S. SARS-CoV-2 antigen tests predict infectivity based on viral culture: Comparison of antigen, PCR viral load, and viral culture testing on a large sample cohort. Clin Microbiol Infect. 2023;29(1):94-100.
[Crossref] [Google Scholar] [PubMed]
- Hodcroft EB. CoVariants: SARS-CoV-2 mutations and variants of interest. 2021.
- Morgan A. COVIRAL webapp back-end. 2023.
- Morgan A. COVIRAL webapp front-end. 2023.
- Lamb YN. Remdesivir: First approval. Drugs. 2020;80(13):1355-63.
[Crossref] [Google Scholar] [PubMed]
- Ahmed A, Song Y, Wadhera RK. Racial/ethnic disparities in delaying or not receiving medical care during the COVID-19 pandemic. J Gen Intern Med. 2022;37(5):1341-1343.
[Crossref] [Google Scholar] [PubMed]
- Watson C. Rise of the preprint: How rapid data sharing during COVID-19 has changed science forever. Nat Med. 2022;28(1):2-5.
[Crossref] [Google Scholar] [PubMed]
- Callahan CJ, Lee R, Zulauf KE. Open development and clinical validation of multiple 3D-printed nasopharyngeal collection swabs: Rapid resolution of a critical COVID-19 testing bottleneck. J Clin Microbiol. 2020;58(8):e00876-e00920.
[Crossref] [Google Scholar] [PubMed]
- Arnaout RA. Cooperation under Pressure: Lessons from the COVID-19 Swab Crisis. J Clin Microbiol. 2021;59(10):e01239-21.
[Crossref] [Google Scholar] [PubMed]
- Tse EG, Klug DM, Todd MH. Open science approaches to COVID-19. F1000Res. 2020;9:1043.
[Crossref] [Google Scholar] [PubMed]
- Perillat L, Baigrie BS. COVID-19 and the generation of novel scientific knowledge: Evidence-based decisions and data sharing. J Eval Clin Pract. 2021;27:708-15.
[Crossref] [Google Scholar] [PubMed]
- Gardener AD, Hick EJ, Jacklin C. Open science and conflict of interest policies of medical and health sciences journals before and during the COVID-19 pandemic: A repeat cross-sectional study: Open science policies of medical journals. JRSM Open. 2022;13(11):20542704221132139.
[Crossref] [Google Scholar] [PubMed]
- Besançon L, Peiffer-Smadja N, Segalas C. Open science saves lives: lessons from the COVID-19 pandemic. BMC Med Res Methodol. 2021;21:117.
[Crossref] [Google Scholar] [PubMed]
- Wood A, Denholm R, Hollings S. Linked electronic health records for research on a nationwide cohort of more than 54 million people in England: data resource. BMJ. 2021;373:n826.
[Crossref] [Google Scholar] [PubMed]
- Rao SN, Manissero D, Steele VR. A systematic review of the clinical utility of cycle threshold values in the context of COVID-19. Infect Dis Ther. 2020;9(3):573-586.
[Crossref] [Google Scholar] [PubMed]
- Satlin MJ, Chen L, Gomez-Simmonds A. Impact of a rapid molecular test for Klebsiella pneumoniae Carbapenemase and Ceftazidime-Avibactam use on outcomes after bacteremia caused by carbapenem-resistant Enterobacterales. Clin Infect Dis. 2022;75:2066-2075.
[Crossref] [Google Scholar] [PubMed]
- Savela ES, Viloria Winnett A, Romano AE. Quantitative SARS-CoV-2 viral-load curves in paired saliva samples and nasal swabs inform appropriate respiratory sampling site and analytical test sensitivity required for earliest viral detection. J Clin Microbiol. 2022;60(2):e0178521.
[Crossref] [Google Scholar] [PubMed]
- Viloria Winnett A, Porter MK, Romano AE. Morning SARS-CoV-2 testing yields better detection of infection due to higher viral loads in saliva and nasal swabs upon waking. Microbiol Spectr. 2022;10(6):e0387322.
[Crossref] [Google Scholar] [PubMed]
- Brummer LE, Katzenschlager S, Gaeddert M. Accuracy of novel antigen rapid diagnostics for SARS-CoV-2: A living systematic review and meta-analysis. PLoS Med. 2021;18(8):e1003735.
[Crossref] [Google Scholar] [PubMed]
- Lee RA, Herigon JC, Benedetti A. Performance of saliva, oropharyngeal swabs, and nasal swabs for SARS-CoV-2 molecular detection: A systematic review and meta-analysis. J Clin Microbiol. 2021;59(5):e02881-e02920.
[Crossref] [Google Scholar] [PubMed]
- El Emam K. Guide to the de-Identification of personal health information. Boca Raton, United States: Auerbach Publishers, Incorporated 2013.
- El Emam K, Rodgers S, Malin B. Anonymising and sharing individual patient data. BMJ. 2015;350:h1139.
[Crossref] [Google Scholar] [PubMed]
Citation: Morgan A, Contreras E, Yasuda M, Dutta S, Hamel D, Shankar T, et al. (2024) The Coviral Portal: Multi-Cohort Viral Loads and Antigen- Test Virtual Trials for COVID-19. J Vaccines Vaccin. S25:001.
Copyright: © 2024 Morgan A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

