Indexed In
- Open J Gate
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer
Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2023) Volume 12, Issue 2
Phylogeny Analysis Reveals the Circulation of Three Geographical Lineages of Rabies Virus in Azerbaijan
Chichak Aliyeva1*, Tamilla Aliyeva1, Khalid Bayramov2, Shalala Zeynalova3, Kadir Yesilbag4, Fahrettin Ozcan5 and Bahtiyar Yilmaz52Department of Animal Health, Azerbaijan State University, Baku, Azerbaijan
3Department of Animal Health, Azerbaijan Veterinary Scientific Institute, Baku, Azerbaijan
4Department of Animal Health, Uludag University, Bursa, Turkey
5Department of Biotechnology, Letgen Biotechnology, Izmir, Turkey
Received: 15-Apr-2023, Manuscript No. CMO-23-21049; Editor assigned: 17-Apr-2023, Pre QC No. CMO-23-21049(PQ); Reviewed: 03-May-2023, QC No. CMO-23-21049; Revised: 10-May-2023, Manuscript No. CMO-23-21049(R); Published: 17-May-2023, DOI: 10.35248/2327-5073.23.12.333
Abstract
Due to the fact that the rabies virus causes lethal infections in humans and animals, it is still considered a hazardous disease. Azerbaijan and surrounding territories are described among the endemic regions for rabies. Geographical conditions allow easy transitions for wild animals from the borders and affect the endemic situations and existence of the rabies virus genotypes in a defined region. Thus the follow-up studies for the current epidemiological situation in the field are crucial.
This study aimed to molecular characterization of the recent field strains of the rabies virus circulating in Azerbaijan between 2018 and 2021. A total of 180 samples out of 238 submitted were found positive for rabies by immunofluorescence assay and real-time Reverse Transcription Polymerase Chain Reaction (RT-PCR) during the study period. Brain samples obtained from 13 infected animals (3 cattle, 3 jackals, 3 dogs, 1 cat, 2 horses and 1 fox) were submitted for sequence analysis on the basis of the N gene. Eleven out of 13 sequences were found in the Central Asian clusters CA4 and CA2 while the remaining 2 sequences were from the genetic cluster Middle East ME1. Despite a high level but not complete nucleotide identity, a phylogenetic relationship between the rabies virus spread in Azerbaijan and in the neighbouring countries (Turkey, Georgia, Iran, as well as Kazakhstan) was demonstrated. The results confirm that the strains spreading in neighbouring countries should be considered in the epidemiology and prevention of rabies.
Keywords
Rabies virus; Neurological disorders; Lyssavirus; Reverse Transcription Polymerase Chain Reaction (RT- PCR)
Abbreviations
N: Nucleoprotein; P: Phosphoprotein; M: Matrix proteins; G: Glycoprotein; GT: Genotypes; ICTV: International Committee on Taxonomy of Viruses; WOAH: World Organization for Animal Health; WHO: World Health Organization; FAO: Food and Agriculture Organization of the United Nations; RT-PCR: Reverse Transcription Polymerase Chain Reaction; ME: Middle East; IFAT: Immunofluorescence Antibody; CA: Central Asian
Introduction
Rabies which is a deadly zoonotic disease of human kind and warm-blooded animals, leads to severe neurological disorders and exists in many territories all around the world [1]. The infection causes about 59000 deaths per year, most are in the developing countries and the disease control attempts cost about 8.6 billion USD per year [2].
Rabies viruses consist of one molecule of genomic RNA (11-12 kb long, non-fragmented) and five structural proteins namely Nucleoprotein (N), Phosphoprotein (P), Matrix proteins (M), Glycoprotein (G) and RNA polymerase (L) [3]. Helically wrapped nucleocapsids are surrounded by lipid membrane with glycoprotein peplomers. For seven Genotypes (GT) of the rabies virus, full studies on the genomic diversity of lyssaviruses have been published using genome sequences (GT1-Rabies Virus, GT2-Lagos bat virus, GT3- Mokola virus, GT4-Duvenhage virus, GT5-European bat lyssavirus type 1, GT6-European bat lyssavirus type 2, GT7-Australian bat lyssavirus) [4]. All genomes have the same structural organization, despite their varying lengths of 11918 nucleotides (GT7) and 12016 nt. (GT2). The estimated size of the coding regions is similar across genotypes.
The agent, Rabies virus is classified in the Lyssavirus genus of the family Rhabdoviridae. Based on delimitation criteria such as antigenic samples reacting with anti-nucleocapsid monoclonal antibody panels and genetic distance, lyssaviruses are classified into to date, 17 species of lyssavirus have been officially recognized by the International Committee on Taxonomy of Viruses (ICTV) [5]. Lyssavirus species are divided into two phylogroups. The antigenic and genetic profile of the virus also allows for separation within phylogroups [6-8]. Phlyogroup-1 includes Rabies lyssavirus, Duvenhage lyssavirus, European bat Lyssaviruses types 1 and 2, Bokeloh bat Lyssavirus and Australian bat Lyssavirus. In addition, Aravan lyssavirus, Khujand lyssavirus and Irkut lyssavirus are also members of phylogroup-1. Phylogroup-2 includes Lagos bat Lyssavirus, Mokola lyssavirus and Shimoni bat Lyssavirus.
Despite conducting various scientific researches on rabies and vaccination among animals on a large scale, this disease remains relevant and dangerous in many countries. Asia, Africa, and the Americas are the most affected regions by rabies among people around the world. In countries located in Asia, approximately 31,000 people lose their lives due to rabies every year [9]. Among these countries, India has the highest number (20,000) of human deaths due to rabies annually [10].
According to a risk potential assessment for rabies, the countries were divided into 3 groups as risk-free, medium-risk and highrisk. According to the evaluations, Azerbaijan, Turkey, Armenia, Georgia, Iran and Russia, as well as many other countries, are included in the high-risk category [11].
Presence of the infection has long been demonstrated in Azerbaijan territory where is an endemic area both for urban and wild life rabies infections [12]. Free-roaming dogs in residential areas are accepted as the leading cause for the human infection in the country while the basins for the river Kura and river Araz as well as the forest massifs in the interior and the border lands are considered the main risk areas. Due to the geographical circumstances the control of country borders is extremely difficult in the Caucasian region, thus allowing free circulation of wild life animals. Despite the mentioned risk level, few studies have been conducted in the country, so far.
Control of rabies is one of the main goals for the WHO and WOAH. In 2015, The international organizations, i.e. the World Organization for Animal Health (WOAH), the World Health Organization (WHO), and the Food and Agriculture Organization of the United Nations (FAO), developed a strategic plan to eliminate dog mediated human rabies by 2030 [13,14]. One of the valuable data for control of the infection is to epidemiological follow up for the field infections and characterization of the circulating strains. Thus, to point out the current situation and to create additional data for country strategic plan, the aim of this study was to molecular characterization of the rabies virus field strains recently obtained from wild and domestic animals in Azerbaijan.
Materials and Methods
Tested population and samples
Studied animal population in the present research is selected among the samples submitted to Central Veterinary Laboratory, Azerbaijan Food Safety Institute, Baku during a period of 4 years (2018-2021). During this four year period samples from total of 238 suspected animals were submitted and 180 animals were tested positive for rabies (68 in 2018, 63 in 2019, 29 in 2020 and 20 in 2021). Distribution and the characteristics of the tested samples are summarized (Table 1). Eight of the rabies positive samples were from wild animals while the remaining originated from domestic animals including dogs (n=102), cats (n=7), cattle (n=54), sheep and goats (n=6) and horses (n=3). All the samples were handled according to biosafety rules and tested by internationally approved methods including Immunofluorescence Antibody assay (IFAT) and Reverse Transcription Polymerase Chain Reaction (RT-PCR). Among the rabies positive samples 13 were selected for sequencing and phylogeny analyses.
| Years | Number of samples | Total | Positives (%) | |||||
|---|---|---|---|---|---|---|---|---|
| Dog | Cat | Cattle | Horse | Sheep and goats | Wild animals | |||
| 2018 | 40 | 2 | 21 | 0 | 3 | 2 | 78 | 68 (87.17%) |
| 2019 | 39 | 4 | 14 | 2 | 1 | 3 | 80 | 63 (78.75%) |
| 2020 | 14 | 1 | 10 | 1 | 1 | 2 | 49 | 29 (59.18%) |
| 2021 | 9 | 0 | 9 | 0 | 1 | 1 | 31 | 20 (64.51%) |
| Total (2018-2021) | 102 | 7 | 54 | 3 | 6 | 8 | 238 | 180 (75.63%) |
Table 1: Distribution of the samples submitted for rabies diagnosis during the period of 2018-2021.
Immunofluorescence assay
In order to carry out this study, brain tissues were dissected pathologically and tissue samples of at least 2 cm3 were collected from the cortex, cerebellum and Ammon horn regions of the brain (5-6 smears). FITC Anti-Rabies Monoclonal Globulin (Fujirebio Diagnostics, Inc. America) was used to stain brain samples. By combining fluorochrome-labelled antibodies with the nucleocapsid of the rabies virus, this method produced greenemerald illumination during fluorescence microscopy (Figure 1).
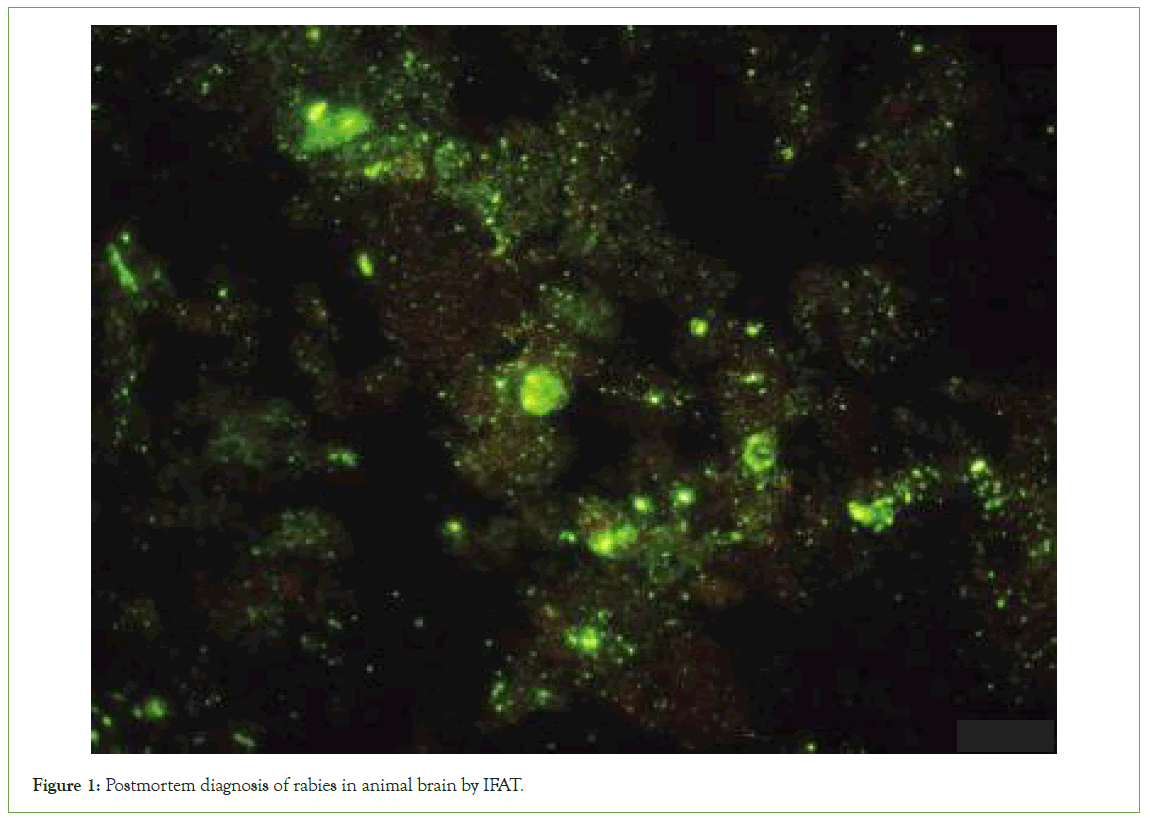
Figure 1: Postmortem diagnosis of rabies in animal brain by IFAT.
Reverse Transcription Polymerase Chain Reaction (RTPCR)
The amplification of the full nucleoprotein gene was performed by a one-step real time RT-PCR (rt RT-PCR) with specific rabies primers designed to specifically amplify the rabies virus genome, followed by a second round of PCR with nested primers for typing. Following the manufacturer’s instructions, using the RT PCR primers N165-146 and JW12 provided a final master mix (QuantiTect probe RT-PCR Mater mix, Qiagen, France) prepared in a single tube per sample. Rt RT-PCR was performed in a final volume of 25 μL with 2 μL of viral RNA and 23 μL of master mix. The PCR thermal profile consisted of; one cycle at 500°C for 30 minutes, one cycle of 950°C for 15 minutes followed by 45 cycles of 950°C for 30 sec, 550°C for 30 sec and 720°C for 30 sec.
After getting the positive result in rt RT-PCR, the samples were subjected to a gel based nested RT-PCR in which the entire N gene amplification (1353-bp) was performed as follows; 5 μL of extracted RNA was mixed with 0.6 μl of One Step RT-PCR Enzyme Mix (Qiagen, France), 0.6 μl of dNTP 10 mM and 10 pmol each of specific rabies primers JW12 and PVN8. In the second round of PCR; 2 μL of PCR products from 1st round was re-amplified in a final volume of 20 μl using 1.25 U of Platinum Taq DNA polymerase (Invitrogen, France) and 2.5 pmol each of sequencing primers M13-JW12 and M13-PVN8bis. The PCR conditions were as follows; initial denaturation for 2 minutes at 94°C, followed by 45 cycles with denaturation for 30 seconds at 94°C, annealing for 30 seconds at 48°C and extension for 2 minutes at 72°C, and a final extension for 7 minutes at 72°C. By nested RT-PCR, the Nucleoprotein gene of the viral RNA was partially amplified. RTPCR and round Polymerase Chain Reactions (nested PCR) were performed with pan-Lyssavirus primers described by Heaton et al., yielding a PCR product of 606 bp.
Phylogeny analysis
Selected 13 samples positive both in IFAT and RT-PCR were subjected to nucleotide sequencing (Table 2). The selection of the samples for sequencing was based on several important considerations. As such, attention was given to their full coverage of the country by region, as well as their diversity in species and wild animals (Figure 2). The cDNA samples were prepared from sample RNAs by a commercial kit (Grisp Xpert One-Step RT-PCR Kit) and sent for sequence analysis (Letgen Biotechnology Laboratory, Izmir, Turkiye).
| Sample ID | Species of origin | Location | Sampling year |
|---|---|---|---|
| Azer 1 | Dog | Baku | 2021 |
| Azer 2 | Jackal | Agdash | 2019 |
| Azer 3 | Cattle | Agstafa | 2020 |
| Azer 4 | Jackal | Lerik | 2019 |
| Azer 5 | Jackal | Lachin | 2020 |
| Azer 6 | Dog | Astara | 2019 |
| Azer 7 | Yearling cattle | Goychay | 2019 |
| Azer 8 | Dog | Salyan | 2021 |
| Azer 9 | Horse | Fuzuli | 2019 |
| Azer 10 | Fox | Gandja | 2021 |
| Azer 11 | Horse | Tovuz | 2020 |
| Azer 12 | Cattle | Qakh | 2019 |
| Azer 13 | Cat | Sheki | 2021 |
Table 2: Characteristics of the field samples selected for phylogeny analysis.
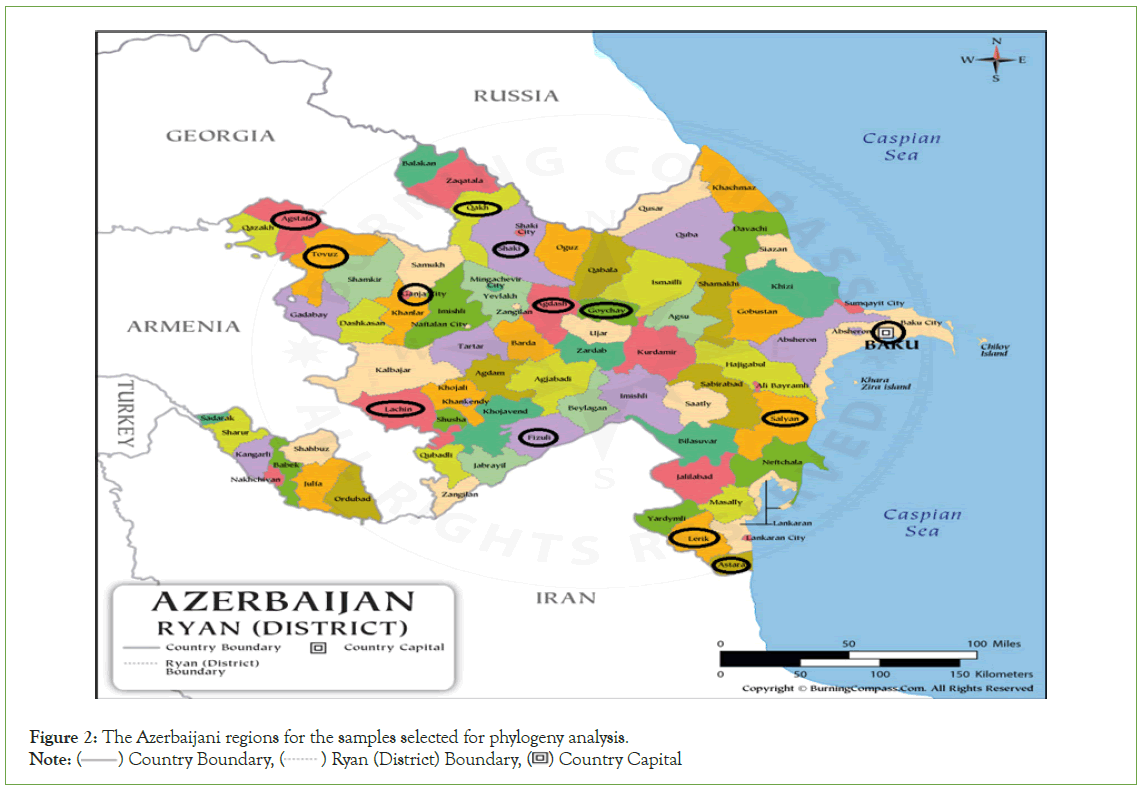
Figure 2: The Azerbaijani regions for the samples selected for phylogeny analysis.
Note: ( ) Country Boundary, (
) Country Boundary, ( ) Ryan (District) Boundary, (
) Ryan (District) Boundary, ( ) Country Capital
) Country Capital
The evolutionary history was inferred using the Neighbor- Joining method. The optimal tree with the sum of branch length=1.85082495 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Tamura-Nei method and are in the units of the number of base substitutions per site. This analysis involved 13 nucleotide sequences. Evolutionary analyses were conducted in MEGA X where the reference sequences were obtained from GenBank databases (Figure 3).
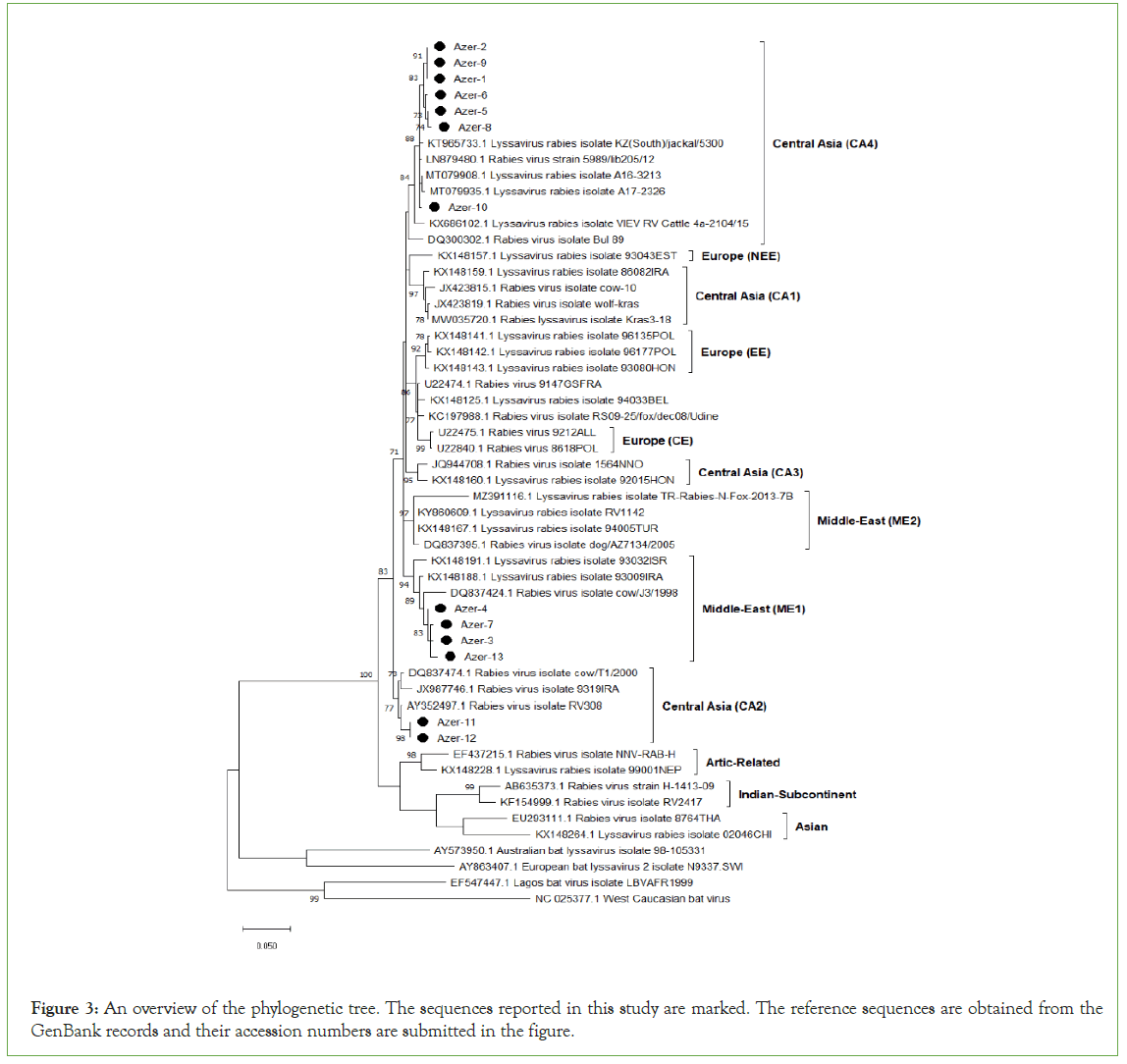
Figure 3: An overview of the phylogenetic tree. The sequences reported in this study are marked. The reference sequences are obtained from the GenBank records and their accession numbers are submitted in the figure.
Results
Screening for rabies infection
Rabies virus screening performed both by IFAT and PCR created the consistent results (Table 3). According to that results, 180 (75.6%) out of 238 samples found positive for rabies. Positivity ratios for the years were as 87.17%, 78.75%, 59.18% and 64.51%, respectively.
| Years | Number of samples | |||
|---|---|---|---|---|
| Tested | Positives by IFAT | Positives by PCR | Positives (%) | |
| 2018 | 78 | 68 | 68 | 87.17% |
| 2019 | 80 | 63 | 63 | 78.75% |
| 2020 | 49 | 29 | 29 | 59.18% |
| 2021 | 31 | 20 | 20 | 64.51% |
| Total (2018-2021) | 238 | 180 | 180 | 75.63% |
Table 3: Rabies screening results for a four years’ period.
Phylogeny analysis
Total of 13 rabies virus sequences from Azerbaijan territories were subjected to phylogeny analysis (Table 4). Those of viral strains were detected to be originated from 3 different genetic clusters namely Central Asian CA2, Central Asian CA4 and the Middle Eastern ME1. Most of the sequences are from the cluster Central Asian CA4 while there are only 2 sequences from the cluster Middle East ME1.
| The new sequence (this study) | Defined cluster | Similar sequence (GenBank Acc. No) | Origine | Nucleotide identity (%) | Collection date |
|---|---|---|---|---|---|
| Azer-1 | CA4 | LN879480.1* | Azerbaijan | 98,98% | 2002 |
| MW055108.1* | Georgia | 98.98% | 2016 | ||
| KT965733.1 | South Kazakhstan | 98.76% | 2014 | ||
| KX148166.1 | Turkiye | 96.73% | 1993 | ||
| Azer-3 | ME1 | MK760742.1* | Iran | 99.19% | 2014 |
| KJ081443.1 | Turkiye | 97.96% | 2013 | ||
| Azer-4 | ME1 | MK760742.1* | Iran | 99.8% | 2014 |
| DQ837466.1 | Israel | 98.58% | 2002 | ||
| KJ081443.1 | Turkiye | 98.57% | 2013 | ||
| Azer-5 | CA4 | LN879480.1* | Azerbaijan | 98,99% | 2002 |
| MW177597.1 | Azerbaijan | 96.15% | 2012 | ||
| OL515137.1 | Romania | 96.15% | 2012 | ||
| OM542196.1 | Poland | 96.15% | 2001 | ||
| Azer-8 | CA4 | LN879480.1* | Azerbaijan | 98,58% | 2002 |
| KT965733.1 | South Kazakhstan | 98.34% | 2014 | ||
| KX148166.1 | Turkiye | 95.94% | 1993 | ||
| Azer-10 | CA4 | LN879480.1* | Azerbaijan | 99.39% | 2002 |
| KT965733.1 | South Kazakhstan | 99.17% | 2014 | ||
| MW035720.1 | Russia | 96.35% | 2018 | ||
| Azer-11 | CA2 | MT079950.1* | Georgia | 99.8% | 2016 |
| LN879480.1 | Azerbaijan | 96.15% | 2002 | ||
| KY002889.1 | Dagestan (Rus) | 99.34% | 2008 | ||
| KX148167.1 | Turkiye | 96.15% | 1993 |
Note: (*) The highest identity in GenBank nucleotide records.
Table 4: Percentile nucleotide identity for selected Azerbaijani rabies virus partial N gene sequences.
Discussion
Azerbaijan and the surrounding Caucasian regions are described as the land of endemic rabies circulation [15]. There are strict border transitions that allow the roaming of wild animals and of course the rabies virus field strains among the countries. Thus, the endemicity of the field strains and genetic clusters in a defined region is based on a dynamic situation. The main goal of the current study is to enlighten the current epidemiological situation regarding the genetic composition of rabies viruses circulated both in the site of urban and wildlife in Azerbaijan territories.
During the 4 years study total of 238 suspected samples from wild and domestic animal from different parts of Azerbaijan have been examined, and of which 180 were found positive for rabies virus infection (Table 3). All the 180 samples were positive both by RTPCR and IF assays confirm the best compatibility of two assays for routine diagnosis in clinical samples. Although the detected positivity rates ranged between 59.18% to 87.17% number of positive samples has been gradually decreased from 68 cases to 20 cases during the years 2018 to 2021. This decrease possibly related to control measure applied in the country [16].
A total of 13 field sample sequences were examined in the present study. Of these, nine samples were identified to be from the cluster Central Asia and found that two different phylogenetic subclades (CA4 and CA2) are currently circulating in the region. CA2 is the one previously described by Troupin et al [17]. Troupin et al. divides the Central Asia clade into three sub-clades. Viruses found in CA1 originated in China, Iran, and Russia; viruses found in CA2 originated in Iraq, Iran, Turkey and Russia; and a cluster of viruses (called CA3) originated in China, Iran, and Russia. CA4 originated in South Kazakhstan, Turkey, Georgia, Romania, Poland and Russia. Remaining sequences were identified in the subclades ME1. Appearance of more than one genetic subclades of rabies virus in the same region is also case for various territories [18].
The phylogenetic analysis is based on the comparison of the partial N gene sequences between 13 Azerbaijani sequences of the study. Thirteen referenced Azerbaijani sequences and 61 referenced Lyssavirus sequences which showed that viruses from dogs predominate in the lineage CA4. According the obtained data 3 dogs, 2 jackals, 1 fox, and 1 horse constituted the lineage CA4. The phylogenetic study among seven isolated from Azerbaijan and referenced Azeri sequences showed the existence of at least two lineages occurring in the country, with a predominance of dogs in the subclade CA4. Involvement of the both of samples from wild and domestic animals in the same clade and 100% homology in the partial N gene sequences of 2 strains (Azer-1 and Azer-2) may indicate the active transmission between wild life and domestic animals.
Conclusion
Based on the genetic analyses conducted within the framework of these studies, the rabies virus group currently circulating in Azerbaijan is a cosmopolitan strain related to dogs. Three sequence results isolated from the Kakheti region of Georgia in 2018 were not identical to other group results and were independently combined. Because the Balakan region of Azerbaijan borders the Kakheti region, the sequence results of the rabies virus spread in those areas were similar, which may be related to the nomadic lifestyle of the population living in those regions. This virus can be spread in the area by animal husbandry animals moving around and wild animals biting productive animals. It appears that several different strains of rabies are circulating simultaneously in Azerbaijan due to its geographical location within the Caucasus region.
Acknowledgement
Authors would like to express their sincere gratitude to Azerbaijan Food Safety Institute for the support provided during the study. We also thank to Dr. Eda Baldan Toker from Bursa Uludag University for her help in phylogeny analysis.
References
- H. Wilde, Tepsumethanon V, in international encyclopedia of public health (2008) extent of the disease and routes of transmission.
- Hampson K, Coudeville L, Lembo T, Sambo M, Kieffer A, Attlan M, et al. Estimating the global burden of endemic canine rabies. PLoS Negl Trop Dis. 2015; 9(4):e0003709.
- King AM, Lefkowitz E, Adams MJ, Carstens EB. Virus taxonomy: Ninth report of the international committee on taxonomy of viruses. Elsevier. 2011.
- Badrane H, Tordo N. Host switching in lyssavirus history from the chiroptera to the carnivora orders. J Virol. 2001; 75(17):8096-8104.
- Hu SC, Hsu CL, Lee F, Tu YC, Chen YW, Chang JC, et al. Novel bat lyssaviruses identified by nationwide passive surveillance in Taiwan, 2018–2021. Viruses. 2022; 14(7):1562.
- White DO, Fenner F.J. Medical Virology. 4th Edition, Academic Press, San Diego. 1994; 475-488.
- Delmas O, Holmes EC, Talbi C, Larrous F, Dacheux L, Bouchier C, et al. Genomic diversity and evolution of the lyssaviruses. PLoS One. 2008;3(4):e2057.
- Rabies Information System of the WHO. 2023.
- Rabies in the South-East Asia Region. 2022.
- Rabies in India. 2015.
- UK G. Rabies risks in terrestrial animals by country. 2021.
- Zeynalova S. Molecular genetic analysis of the rabies virus genome isolated in Azerbaijan. Khazar J Sci Technol. 2020; (4)2: 1-37.
- Minghui R, Stone M, Semedo MH, Nel L. New global strategic plan to eliminate dog-mediated rabies by 2030. Lancet Glob Health. 2018;6(8):e828-829.
- World Health Organization. Zero by 30: The global strategic plan to end human deaths from dog-mediated rabies by 2030. 2018; 1-47.
- Zeynalova S, Shikhiyev M, Aliyeva T, Ismayilova R, Wise E, Abdullayev R, et al. Epidemiological characteristics of human and animal rabies in Azerbaijan. Zoonoses Public Health. 2015; 62(2):111-118.
- Hasanov E. Rabies control measures in Azerbaijan: Survey of stray dog population in Baku. J Agric Vet. 2021; 5(2): 70-77.
- Troupin C, Dacheux L, Tanguy M, Sabeta C, Blanc H, Bouchier C, et al. Large-scale phylogenomic analysis reveals the complex evolutionary history of rabies virus in multiple carnivore hosts. PLoS Pathog. 2016; 12(12):e1006041.
- Mey C, Metlin A, Duong V, Ong S, In S, Horwood PF, et al. Evidence of two distinct phylogenetic lineages of dog rabies virus circulating in Cambodia. Infect Genet Evol. 2016;38:55-61.
Citation: Aliyeva C, Aliyeva T, Bayramov K, Zeynalova S, Yesilbag k, Ozcan F, et al. (2023) Phylogeny Analysis Reveals the Circulation of Three Geographical Lineages of Rabies Virus in Azerbaijan. Clin Microbiol. 12: 333.
Copyright: © 2023 Aliyeva C, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
