Indexed In
- Open J Gate
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer
Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2025) Volume 14, Issue 2
Genotyping of Soil and Clinical Isolates of Burkholderia Species in Myanmar
Nay Myo Aung1,3*, Khine Khine Su3, Narisara Chantratita2 and Chanwit Tribuddharat12Department of Microbiology and Immunology, Mahidol University, Bangkok, Thailand
3Department of Microbiology, Defense Services Medical Academy, Yangon, Myanmar
Received: 09-Feb-2024, Manuscript No. CMO-24-24885; Editor assigned: 12-Feb-2024, Pre QC No. CMO-24-24885 (PQ); Reviewed: 26-Feb-2024, QC No. CMO-24-24885; Revised: 03-Apr-2025, Manuscript No. CMO-24-24885 (R); Published: 11-Apr-2025, DOI: 10.35248/2327-5073.25.14.429
Abstract
Burkholderia species are known as medically necessary pathogens not only in a clinical setting but environmental sectors. Cost-effective ways of molecular genotyping among Burkholderia species in Myanmar were found to be limited and low awareness. In this study, 21 clinical and 29 soil isolates were collected and performed to discriminate Burkholderia species. Two genotyping methods were performed, recA, 16S rRNA amplification and sequencing. Phylogenetic trees were constructed using sequences of recA and 16S rRNA genes. Twenty-nine soil isolates resulted as 5 Burkholderia pseudomallei, 5 Burkholderia cepacia, 13 Burkholderia thailandensis, 4 Burkholderia mutivorans and 2 Burkholderia cenocepacia. Sixteen clinical and 5 soil isolates of Burkholderia pseudomallei were taken and assessed 16S rRNA type. RecA sequencing was grouped into 5 clades, while 16S rRNA sequencing showed two. The more type ability and discriminatory power of recA were found than 16S rRNA sequencing among Burkholderia species. The recA sequencing showed a greater discriminatory power and type ability, suggesting that it may be a more reliable and helpful tool for identifying and classifying Burkholderia species. Therefore, recA sequencing can be considered a better alternative to 16S rRNA sequencing for the molecular characterization of Burkholderia species in Myanmar.
Keywords
Burkholderia species; Genotyping; recA gene; Myanmar
Introduction
Burkholderia species are found in various environments, including soil, water and plants [1]. These bacteria are capable of causing severe diseases in humans, animals and plants and their clinical importance is well-recognized. Burkholderia species have been implicated in various infections, including pneumonia, urinary tract infections, sepsis and wound infections [2]. In particular, B. pseudomallei area significant cause of morbidity and mortality in regions where it is endemic, including Southeast Asia and northern Australia [3].
Burkholderia species exhibit high genomic diversity, making molecular genotyping crucial for their identification and classification [4]. Molecular genotyping methods have been shown to have higher resolution and discriminatory power than traditional phenotypic methods, making them a more reliable tool for identifying and classifying Burkholderia species [5]. Two commonly used molecular genotyping methods for Burkholderia species are recA and 16S rRNA gene sequencing.
The recA gene encodes a recombinase enzyme involved in DNA repair and recombination. It is highly conserved among bacterial species and has been used as a target for molecular genotyping and phylogenetic analysis of Burkholderia species [6]. In contrast, the 16S rRNA gene is highly conserved in all bacterial species and is involved in translating mRNA to protein. It has been widely used as a target for molecular identification and classification of bacterial species, including Burkholderia species [7].
Several studies have compared the usefulness of recA and 16S rRNA sequencing for identifying and classifying Burkholderia species. One study reported that recA sequencing had a higher discriminatory power and was more effective in identifying B. pseudomallei isolates than 16S rRNA sequencing [8,9]. Another study found that recA sequencing could differentiate closely related Burkholderia species, while 16S rRNA sequencing showed limited discriminatory power [10].
In addition to recA and 16S rRNA sequencing, other molecular genotyping methods have been used to identify and classify Burkholderia species. These methods include Multilocus Sequence Typing (MLST), which targets several housekeeping genes and Whole-Genome Sequencing (WGS), which provides a high-resolution view of the genetic diversity of Burkholderia species [11].
In Myanmar, limited information is available on the prevalence, diversity and clinical significance of Burkholderia species. A previous study reported the isolation of B. pseudomallei from soil and water samples in Myanmar [12,13]. However, there is a need to expand the knowledge of the prevalence of other Burkholderia species in the country and to develop cost-effective and reliable molecular genotyping methods for their identification and classification according to local facilities in Myanmar [14].
Molecular genotyping methods are essential for accurately identifying and classifying Burkholderia species. However, rapid screening of molecular genotyping among Burkholderia species in Myanmar was found to be a limitation and low awareness. The current diagnostic methods in Myanmar for identifying Burkholderia species are limited to phenotypic methods, which have limitations in their sensitivity and specificity. Furthermore, more than 16S rRNA sequencing alone to identify Burkholderia species may be required due to the high diversity of Burkholderia species. The development of cost-effective and reliable molecular genotyping methods can contribute to the prevention and control of infections caused by these medically necessary pathogens in Myanmar.
Several molecular genotyping methods have been used to identify and classify Burkholderia species in different settings. WGS has also been increasingly used for identifying and classifying Burkholderia species, providing a high-resolution view of the genetic diversity and evolution of these organisms [15,16]. However, these methods can be resource-intensive and costly, limiting their use in resource-limited settings such as Myanmar. This study highlighted the simple discrimination of Burkholderia species among clinical and soil isolates by using a local affordable and properly facilitated laboratory in Myanmar.
Materials and Methods
Bacterial strain collection
Sixteen clinical isolates of Burkholderia pseudomallei were taken and assessed for 2 sequencing assays in a previous study [17]. We collected 29 soil isolates from a gift of the study of Htun et al. [13]. At first, those colonies of some soil isolates showed positive results in the latex agglutination test, which was bought from Thailand. At the same time, some were found as weakly positive [18]. Next, they were cultured on Ashdown’s agar containing 10 g/L trypticase soy broth (Oxoid), 40 mL/L glycerol (Ultrapure), 5 mg/L 0.1% crystal violet (Sigma-Aldrich), 50 mg/L neutral red, 5 mg/L gentamicin (Gibco, Waltham, MA, USA) and 15 g/L agar. They incubated aerobically at 42°C for 4 days to assess if they could grow. They were kept in trypticase soy broth with 20% glycerol at -20°C.
Ethics review
The Siriraj Institutional Review Board approved the study (SIRB number: 546/2562 (EC1).
Confirmation for B. pseudomallei by 16S rRNA and recA sequencing assay
Bacterial DNA extraction was done using the QIAamp DNA mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocols. For 16S rRNA sequencing, we amplified a 1,488-bp-long fragment covering the whole 16S rRNA gene using the primers F229 (5-CGC AAG CGA AAG TAT CAA GA-3′) and R1908 (5-TTT ACA GCC GAT AAG CGT GAG-3′) according to a previously published article. We sequenced an 869-bp fragment of the recA gene using the previously published primers BUR1, 5-GAT CGA (A/G) AA GCA GTT CGG CAA-3′ and BUR2, 5-TTG TCC TTG CCC TG (A/G) C CG AT-3′. The PCR detection and mixture were described in a previous study [17]. Phylogenetic trees were constructed with each of 16S rRNA and recA sequences using MEGA software and analyzed the discriminatory power and type ability among the above gene sequencing assays.
Results
The 1,488-bp long nucleotide sequences of the whole length of 16S rRNA gene from 21 B. pseudomallei strains were investigated, aligned and compared. Eight different nucleotide positions (positions 157, 249, 651, 851, 968, 1232 and 1274) were found and no gaps were detected [18]. Among 23 16S rRNA genes of B. pseudomallei, 16S rRNA type 1 was probably identified in 15 clinical and 5 soil isolates (86.9%), while 1 clinical isolate (MMBP010) (4.3%) was probably 16S rRNA type 2 (Table 1) [18].
| 16S rRNA | Difference at position | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Type | 75 | 157 | 249 | 651 | 851 | 968 | 1232 | 1274 | 1292 | 1447 | 1450 | 1451 | 1454 | 1468 | No. of isolate |
| 1 | C | G | C | C | C | T | G | C | C | 20 | |||||
| 2 | A | C | 1 | ||||||||||||
| 3 | R | - | |||||||||||||
| 4 | Y | - | |||||||||||||
| 5 | A | Y | - | ||||||||||||
| 6 | R | - | |||||||||||||
| 7 | Y | - | |||||||||||||
| 8 | A | - | |||||||||||||
| 9 | Y | - | |||||||||||||
Table 1: Position of base differences in 16S rRNA type of studied B. pseudomallei.
Among the selected 15 soil isolates which were latex positive, 5 isolates were identified as B. pseudomallei (n=5) using 16S rRNA gene sequencing. In the case of the remaining 10 isolates, 16S rRNA gene sequencing can examine only one isolate (Letpandan_NMBt011) to be B. thailandensis (n=1) (Table 2). In contrast, the remaining 9 soil isolates showed contaminated peaks in the chromatogram of the DNA sequencing. For 14 soil isolates that were latex negative, 13 isolates were confirmed as B. thailandensis, while 1 isolate could not be identified. For 22 clinical Burkholderia isolates, B. pseudomallei (n=19) and B. cepacia (n=3) were identified by 16S rRNA gene sequencing in a previous study [17].
| No (n=29) | Latex agglutination | Strain | Species investigation | |
|---|---|---|---|---|
| 16S rRNA (Whole gene 1488bp)a | recA (820-870 bp)a | |||
| 1 | + | Tontae_NMBP001 | B. pseudomallei | B. pseudomallei |
| 2 | + | Tontae_NMBP002 | B. pseudomallei | B. pseudomallei |
| 3 | + | Tontae_NMBP003 | B. pseudomallei | B. pseudomallei |
| 4 | + | Pathein_NMBP004 | B. pseudomallei | B. pseudomallei |
| 5 | + | Pathein_NMBP005 | B. pseudomallei | B. pseudomallei |
| 6 | - | Nyaung-don_NMBt001 | B. thailandensis | B. thailandensis |
| 7 | - | Kyaung Taung_NMBt002 | B. thailandensis | B. thailandensis |
| 8 | - | Pathein_NMBt003 | B. thailandensis | B. thailandensis |
| 9 | - | Bago_NMBt004 | B. thailandensis | B. thailandensis |
| 10 | - | Tontae_NMBt005 | B. thailandensis | B. thailandensis |
| 11 | - | Bago_NMBt006 | B. thailandensis | B. thailandensis |
| 12 | - | Kyaiklat_NMBt007 | B. thailandensis | B. thailandensis |
| 13 | - | Bago_NMBt008 | B. thailandensis | B. thailandensis |
| 14 | - | Bago_NMBt009 | B. thailandensis | B. thailandensis |
| 15 | - | Letpandan_NMBt010c | B. thailandensis | B. thailandensis |
| 16 | - | Tontae_NMBt011 | B. thailandensis | B. thailandensis |
| 17 | - | Tontae_NMBt012 | B. thailandensis | B. thailandensis |
| 18 | - | Bago_NMBt013 | B. thailandensis | B. thailandensis |
| 19 | + | Mawbi_NMBcc001b | â?? | B. cenocepacia |
| 20 | + | Mawbi_NMBcc002b | â?? | B. cenocepacia |
| 21 | + | Mawbi_NMBcc003b | â?? | B. cenocepacia |
| 22 | - | Kyaiklat_NMBcc004 | â?? | B. cenocepacia |
| 23 | + | Eainme_NMBcc005b | â?? | B. cenocepacia |
| 24 | + | Mada_NMBmu001b | â?? | B. multivorans |
| 25 | + | Mada_NMBmu002b | â?? | B. multivorans |
| 26 | + | Mawbi_NMBmu003b | â?? | B. multivorans |
| 27 | + | Mawbi_NMBmu004b | â?? | B. multivorans |
| 28 | - | Zalon_NMBc001 | â?? | B. cepacia |
| 29 | + | Bogale_NMBc002b | â?? | B.cepacia |
| Note: aThe criteria utilized in this study were as follows: Species level, ≥ 99.0% identity; species-like level, ≥ 98.0% to <99.0% identity; genus level, <98.0% identity, bCross-reactive with mAb-based latex agglutination test, cCross-reactive with mAb-based latex agglutination test and 16S rRNA gene sequencing could not amplify isolates | ||||
Table 2: Species identification by 16S rRNA and recA gene sequencing for soil isolates
A recA gene sequencing could discriminate all 45 soil and clinical isolates in contrast to 16S rRNA sequencing. Overall, a total of 29 soil isolates were investigated and there were B. pseudomallei (n=5), B. cepacia (n=2), B. cenocepacia (n=5), B. multivorans (n=4), and B. thailandensis (n=13). In contrast, recA gene sequencing presented higher discriminatory power than 16S rRNA gene sequencing. For 16 clinical isolates, recA gene sequencing identified B. pseudomallei (n=16) in the previous study [17].
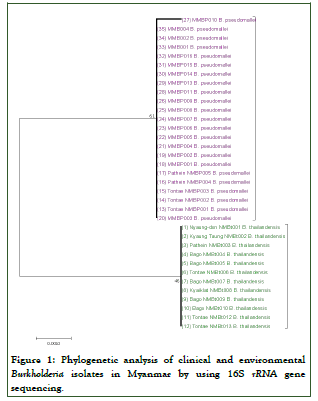
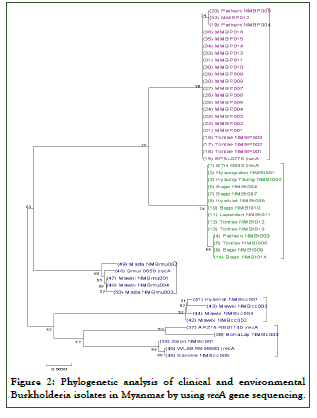
Complete gene sequences of 16S rRNA (n=34) and recA (n=45) were determined in this study and recorded in GenBank with accession nos. CP041221.1 (B. pseudomallei), CP022217.1 (B. thailandensis), HM598390.1 to CP019678.1 (B. cenocepacia), LC487014.1 (B. cepacia) and HM598378.1 (B. multivorans) (Figures 1 and 2).
Figure 1: Phylogenetic analysis of clinical and environmental Burkholderia isolates in Myanmar by using 16S rRNA gene sequencing.
Figure 2: Phylogenetic analysis of clinical and environmental Burkholderia isolates in Myanmar by using recA gene sequencing.
Discussion
Tun Win and colleagues conducted a study in Myanmar to detect soil B. pseudomallei isolates using culture and latex agglutination tests [13]. The research was initiated during the hot season, following standard soil sampling procedures. However, during the wet season in this study, the true B. pseudomallei could rarely be identified, despite the isolates showing positive results in the latex agglutination after 16S rRNA and recA sequencing [19].
In contrast to this approach, this study utilized a latex agglutination test (4B11) purchased from the department of immunology and microbiology at tropical medicine. This test targeted the exopolysaccharide of B. pseudomallei and was used to screen Burkholderia species [20]. Further molecular identification of Burkholderia species, which showed positive agglutination tests, was conducted in the study. Given the resource constraints in Myanmar, the isolates were collected first and, next, performed molecular tests, identifying cross-reactive isolates capable of growing in Ashdown agar and exhibiting agglutination.
Interestingly, this study revealed that the identified strains expressed a B. pseudomallei-like capsular polysaccharide (BTCV), commonly found in Southeast Asian soil and remains nonpathogenic. This finding is consistent with the study conducted by Jirarat et al. The study highlighted the potential of utilizing molecular techniques to identify B. pseudomallei in resourcelimited settings such as Myanmar.
This study proposed a specific identification method for Burkholderia pseudomallei in Myanmar using 16S rRNA gene sequencing [17]. The agreement was found with the previous study by Gee et al., about using 16S rRNA gene sequencing for screening B. pseudomallei. The present study showed that the 16S rRNA gene sequences of Myanmar isolates were consistent with type I and II, revealing the same mutation point locations on a 1488 bp long gene fragment. The 16S rRNA type I incidence was higher in Myanmar isolates (90%) than type II, indicating a potential epidemiological tool for distributing B. pseudomallei.
However, further studies with more B. pseudomallei are needed to prove the representative samples. The long-range primer targeting the whole length of the 16S rRNA gene was used and no amplification of other soil Burkholderia species was observed. The variable region of the 16S rRNA gene was recommended as the target to improve identification accuracy.
Moreover, due to the inability of 16S rRNA gene sequencing to discriminate Burkholderia species from other soil species, recA gene sequencing was performed for identification [8]. This study was the first work in the identification of Burkholderia species in Myanmar. The findings agreed with a previous study in India, where recA gene sequencing showed higher resolution in identifying Burkholderia species than 16S rRNA gene sequencing by analyzing more clades of the phylogenetic tree in this study. Little is known about whether the recA gene of Burkholderia species in Myanmar is cross-reactive with other species, including Bordetella, Pandoraea, Ralstonia or Xanthomonas [8]. The proposed identification methods using 16S rRNA and recA gene sequencing could be used to identify B. pseudomallei and Burkholderia species in Myanmar, respectively. Further studies are needed to strengthen the findings and to investigate crossreactivity with other soil species.
Conclusion
This study showed that the latex agglutination test exhibited problems in accurate identification, screening or culturing among soil isolates. Therefore, additional confirmatory tests were required to isolate true B. pseudomallei in soil; in confirmation of B. pseudomallei, recA sequencing described a higher discriminatory power than 16S rRNA gene sequencing. This work provided the comparative assessment of sequencing among two genes, highlighting the correct choice for sequencing methods with cost-effectiveness in Myanmar.
Acknowledgments
We are grateful to all the laboratory practitioners in Myanmar who collected and stored the leftover samples. We express our gratitude to Mahidol neighboring countries grant's support for this publication. We are obligated to our colleagues working at the Department of Microbiology, Faculty of Medicine Siriraj Hospital, Mahidol University.
Conflict of Interest
None to declare.
References
- Coenye T, Vandamme P. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ Microbiol. 2003;5(9):719-729.
[Crossref] [Google Scholar] [PubMed]
- Tavares M, Kozak M, Balola A, Sa-Correia I. Burkholderia cepacia complex bacteria: A feared contamination risk in water-based pharmaceutical products. Clin Microbiol Rev. 2020;33(3):10-128.
[Crossref] [Google Scholar] [PubMed]
- Limmathurotsakul D, Wongratanacheewin S, Teerawattanasook N, Wongsuvan G, Chaisuksant S, Chetchotisakd P, et al. Increasing incidence of human melioidosis in Northeast Thailand. Am J Trop Med Hyg. 2010;82(6):1113.
[Crossref] [Google Scholar] [PubMed]
- Tuanyok A, Stone JK, Mayo M, Kaestli M, Gruendike J, Georgia S, et al. (2012) The genetic and molecular basis of O-antigenic diversity in Burkholderia pseudomallei lipopolysaccharide. PLoS Negl Trop Dis. 2012;6(1):e1453.
[Crossref] [Google Scholar] [PubMed]
- Sabat AJ, Budimir A, Nashev D, Sa-Leao R, van Dijl JM, Laurent F, et al. Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Euro Surveill. 2013;18(4):20380.
[Crossref] [Google Scholar] [PubMed]
- Mannweiler O, Pinto-Carbo M, Lardi M, Agnoli K, Eberl L. Investigation of Burkholderia cepacia complex methylomes via single-molecule, real-time sequencing and mutant analysis. J Bacteriol. 2021;203(12):10-128.
[Crossref] [Google Scholar] [PubMed]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173(2):697-703.
[Crossref] [Google Scholar] [PubMed]
- Payne GW, Vandamme P, Morgan SH, LiPuma JJ, Coenye T, Weightman AJ, et al. Development of a recA gene-based identification approach for the entire Burkholderia genus. Appl Environ Microbiol. 2005;71(7):3917-3927.
[Crossref] [Google Scholar] [PubMed]
- Chantratita N, Wuthiekanun V, Limmathurotsakul D, Thanwisai A, Chantratita W, Day NP, et al. Prospective clinical evaluation of the accuracy of 16S rRNA real-time PCR assay for the diagnosis of melioidosis. Am J Trop Med Hyg. 2007;77(5):814-817.
[Google Scholar] [PubMed]
- Ginther JL, Mayo M, Warrington SD, Kaestli M, Mullins T, Wagner DM, et al. Identification of Burkholderia pseudomallei near-neighbor species in the Northern Territory of Australia. PLoS Negl Trop Dis. 2015;9(6):e0003892.
[Crossref] [Google Scholar] [PubMed]
- Drevinek P, Mahenthiralingam E. Burkholderia cenocepacia in cystic fibrosis: Epidemiology and molecular mechanisms of virulence. Clin Microbiol Infect. 2010;16(7):821-830.
[Crossref] [Google Scholar] [PubMed]
- Win MM, Ashley EA, Zin KN, Aung MT, Swe MM, Ling CL, et al. Melioidosis in Myanmar. Trop Med Infect Dis. 2018;3(1):28.
[Crossref] [Google Scholar] [PubMed]
- Win TT, Su KK, Than AM, Htut ZM, Pyar KP, Ashley EA, et al. Presence of Burkholderia pseudomallei in the ‘Granary of Myanmar’. Trop Med Infect Dis. 2019;4(1):8.
[Crossref] [Google Scholar] [PubMed]
- Swe MM, Win MM, Cohen J, Phyo AP, Lin HN, Soe K, et al. Geographical distribution of Burkholderia pseudomallei in soil in Myanmar. PLoS Negl Trop Dis. 2021;15(5):e0009372.
[Crossref] [Google Scholar] [PubMed]
- Shafiq M, Ke B, Li X, Zeng M, Yuan Y, He D, et al. Genomic diversity of resistant and virulent factors of Burkholderia pseudomallei clinical strains recovered from Guangdong using whole genome sequencing. Front Microbiol. 2022;13:980525.
[Crossref] [Google Scholar] [PubMed]
- Rachlin A, Mayo M, Webb JR, Kleinecke M, Rigas V, Harrington G, et al. Whole-genome sequencing of Burkholderia pseudomallei from an urban melioidosis hot spot reveals a fine-scale population structure and localised spatial clustering in the environment. Sci Rep. 2020;10(1):5443.
[Crossref] [Google Scholar] [PubMed]
- Aung NM, Su KK, Chantratita N, Tribuddharat C. Workflow for identification of Burkholderia pseudomallei clinical isolates in Myanmar. Jpn J Infect Dis. 2023;76(2):106-112.
[Crossref] [Google Scholar] [PubMed]
- Gee JE, Sacchi CT, Glass MB, de BK, Weyant RS, Levett PN, et al. Use of 16S rRNA gene sequencing for rapid identification and differentiation of Burkholderia pseudomallei and B. mallei. J Clin Microbiol. 2003;41(10):4647-4654.
[Crossref] [Google Scholar] [PubMed]
- Limmathurotsakul D, Dance DA, Wuthiekanun V, Kaestli M, Mayo M, Warner J, et al. Systematic review and consensus guidelines for environmental sampling of Burkholderia pseudomallei. PLoS Negl Trop Dis. 2013;7(3):e2105.
- Anuntagool N, Naigowit P, Petkanchanapong V, Aramsri P, Panichakul T, Sirisinha S. Monoclonal antibody-based rapid identification of Burkholderia pseudomallei in blood culture fluid from patients with community-acquired septicaemia. J Med Microbiol. 2000;49(12):1075-1078.
[Crossref] [Google Scholar] [PubMed]
Citation: Aung NM, Su KK, Chantratita N, Tribuddharat C (2025) Genotyping of Soil and Clinical Isolates of Burkholderia Species in Myanmar. Clin Microbiol. 14:429.
Copyright: © 2024 Aung NM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.