Indexed In
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2025) Volume 9, Issue 3
Detection of Aminoglycoside Resistance Genes in Clinical Isolates of Acinetobacter baumannii
Sumbal Shahzadi*Received: 12-Oct-2023, Manuscript No. JCMS-23-23580; Editor assigned: 16-Oct-2023, Pre QC No. JCMS-23-23580 (PQ); Reviewed: 30-Oct-2023, QC No. JCMS-23-23580; Revised: 08-Apr-2025, Manuscript No. JCMS-23-23580 (R); Published: 15-Apr-2025
Abstract
Acinetobacter baumannii is an emergent nosocomial, gram negative, coccobacilli pathogen that have ability to quickly acquire resistance against multiple antibiotics. Aminoglycoside is class for antibacterial drugs used for pathogenic organism and causes protein synthesis inhibition, irreversible attachment on ribosomal binding site and enzymatic degradation. A. baumannii has acquired resistance against aminoglycosides using multiple genes which produces aminoglycosides modifying enzymes for resistance. The aim of this study was to investigate the prevalence of gene encoding for three different aminoglycoside-modifying enzymes in A. baumannii. 50 strains of A. baumannnii were collected from Mayo Hospital, Services Institute of Medical Sciences and Children Hospital and Institute of Child Health. These strains were characterized by performing biochemical and API test and preceded for antibiotic susceptibility testing. Highest resistance rate was 100% observed for Doxycyclin and Ceftriaxone. Out of 50 drug resistant strains, 38 strains (76%) were found aminoglycosides resistant. After genomic isolation of aminoglycoside resistant strains, PCR was performed to detect aacC1, aadB and aph3 using the specific primers. aacC1 (Acetyletransferases), aadB (“2’-Aminoglycosidenucleotidyltransferase) and aph3 (2’-Aminoglycosidenucleotidyltransferase) were detected with 44%, 34% and 10.5% respectively of drug resistance A. baumannii strains. 24% strains were resistant to aminoglycosides but no resistance gene was detected showing they might involve some other resistance inducing genes. So, there is requirement to further explore other aminoglycoside resistance genes to develop novel drugs.
Keywords
Acinetobacter baumannii; Aminoglycoside resistance; Transmission Electron microscopy
Introduction
Acinetobacter baumannii
Acinetobacter baumannii (A. baumannii) is an aerophilic nonfermenting gram-negative rod-shaped bacterium that is a member of the Moraxellaceace family. These are taxonomically classified as γ-proteobacteria, family Moraxellaceae and Pseudomonadales in order. The significant member of this family, the A. baumannii, has arisen as one of highly bothersome pathogens for the institutions of health care worldwide. A. baumannii cells are about 1.5 μm long, with coccobacillary shape.

Figure 1: Transmission Electron Microscopy (TEM) of A. baumannii on dry surface.
Biochemical characteristics
Genus Acinetobacter consist of some biochemical properties such as catalase positive, oxidase-negative, non-fastidious, non-lactosefermenting, non-motile, citrate positive, and aerobic Gramnegative coccobacilli. Clinical implication of A. baumannii chiefly over last 15 years, has been impelled by its amazing capability to acquire or up-regulate resistance determining factor, creating it one of the entities intimidating recent antibiotic era (Table 1) [1].
| S. no. | Biochemical properties | Results |
|---|---|---|
| 1 | Gram stain | _ |
| 2 | Microscopic shape | Coccobacilli |
| 3 | Lactose fermenting | _ |
| 4 | Oxidase test | _ |
| 5 | Catalase production | + |
| 6 | Coagulase | _ |
| 7 | Urease production | Variable |
| 8 | Motility test | _ |
| 9 | Indole production | _ |
| 10 | Citrate test | + |
Table 1: biochemical properties of A. baumannii.
Transmission
These bacteria usually infect utmost hospitalized patients in vulnerable condition, individuals who are in critical situation with ruptures in the skin integrity and the airway defense. Versatile organism can use a variety of both energy and carbon sources. Performing in synergy with such evolving resistance profile is strange ability of the A. baumannii to stay alive for long time periods all over some hospital surroundings, hence enhancing its capability for the nosocomial spread. These characteristics elucidate the ability of Acinetobacter species to survive in both dry and moist circumstances in a hospital environment, thus contributing in the transmission process. About more than 75% of the hospitalized patients and 2-40 % of the healthy people and be there as carriers of the Acinetobacter specie on their skin A. baumannii can be obtained from water, soil, humans and animals being omnipresent in nature. Acinetobacter spp. are regular residents of the human skin in community but are as well frequently obtained from respiratory tract of the hospitalized patients.
Infection
It has been stated in various reviews seeing back to 1970’s, hospital acquired pneumonia is most common infection affected by A. baumannii still. A. baumannii is now predictable for reasoning of a wide range of extreme nosocomial infections, together with soft tissue and urinary tract infections, skin infections, wound infections, blood stream infections and secondary meningitis. Such nosocomial infections are targeted to seriously ill patients with broad-spectrum antibiotics, those are hospitalized in the Intensive Care Units (ICUs). A. baumannii pneumonia (community acquired) is more severe than the nosocomial pneumonia, that is normally harmful (can cause death in the 8 days of diagnosis) and mortality proportions may be high about 60% [2].
Figure 2: Infections caused by A. baumannii in human.
Symptom
Symptoms of A. baumannii attack includes red and swollen area or wounds if bacteria has attacked by skin. In case of respiratory tract infections there might be cough, pain in chest and trouble during breath. Whereas for feel of burning during urinate while the bacteria has caused the urinary tract infections. Hospitalacquired infections are frequently seen in the critically ill patients those have definite risk factors for emerging an A. baumannii infection contain extended hospital stays, advanced age, immune suppression, presence of comorbid diseases, previous antibiotic use, major trauma or burns, invasive procedures, and presence of mechanical ventilation.
Virulence factors in pathogenesis
Virulence factors which play important role to pathogenesis of A. baumannii, comprises porins, lipopolysaccharides, capsular polysaccharides, phospholipases, metal acquisition systems, protein secretion systems and outer membrane vesicles. Prions take part in adherence, invasion, serum resistance, biofilm formation and persistence. Capsular polysaccharides play role in biofilm formation and growth in serum. Lipopolysaccride and phosphlipases play role in serum resistance survival in tissue infection and immune response. Metal acquisition system irresponsible for persistence and killing of host cell.
Drugs used against A. baumannii
Several classes of drugs are being used against A. baumannii such as β-lactams, aminoglycosides, quinolones, carbapenems, tetracyclines, cephalosporins, lipopeptides and flouroquinolones. But then again bacteria can still get resistant. To avoid it, antibacterial are being used in combination to overcome the clinical effects caused by A. baumannii. This is supposed to increase the coverage. Resistance against the antimicrobial agents is major benefit of A. baumannii in the nosocomial environment. MDR in the bacteria A. baumannii can be resistant against over and above 2 to 5 drug families or classes: The anti-pseudomonal carbapenems (imipenem or meropenem), another antipseudomonal cephalosporins (ceftazidime or cefepime), ampicillin-sulbactam, aminoglycosides (gentamicin, tobramycin or amikacin) lipopeptides (colistin and polymyxin) and fluoroquinolones (ciprofloxacin or levofloxacin). A. baumannii is an adaptable pathogen, with growing consequences in many of the infections that are acquired in hospitals especially amongst the patients of ICU. Antimicrobial resistance is basic reason for the spread of A. baumannii. Antiseptics infection control, antimicrobial therapy, resistance genes, molecular epidemiology take part in the major testing of susceptibility that has active therapeutic potential against the A. baumannii. These might cover all aminoglycosides, fluoroquinolones and beta-lactams (including carbapenems and sub-lactam).
Drug resistance
Acinetobacter specie has developed resistance against nearly all of the antimicrobial agents which are accessible at this time, together with the quinolones, broad-spectrum β-lactams and aminoglycosides. Majority of the strains have got resistance to the cephalosporins, even though resistance against the carbapenems is being increasingly reported. Trouble in destroying these bacteria has permitted them to inhabit niches left unoccupied subsequent extermination of microbes that are highly susceptible. Chief machinery of resistance against the β- lactam in the Acinetobacter species is production of the β- lactamases coded whichever by the plasmid or else by chromosome. Furthermore, short penetrability of outer most membrane of the Acinetobacter, causing from minor pore size of outer-membrane or restricted production or assembly of porin. Along with modifications in the Penicillin Binding Proteins (PBPs) affinity, it has been involved in the Acinetobacter resistance against such antibiotics.
The over-expressing of chromosomal cephalosporinase the AmpC played vital part in creating resistance against the blactam antibiotics. Then again since the late 1970’s growing figures of highly resistant strains have been listed. One of the modes of resistance used by carbapenems and cephalosporins is change in Outer Membrane Proteins (OMPs). Target site alteration is also made by quinolones. Active efflux is used by tetracycline. A. baumannii presents a high level of the resistance against both trimethoprim–sulphamethoxazole and chloramphenicol, however in accordance to genetic basis of the resistance to such compounds in these bacteria little is known.
Aminoglycosides
Aminoglycoside is a bacteriologic and therapeutic class for antibacterial medications of gram negatives which prevent synthesis of protein and comprise as a part of molecule that is amino-modified glycoside. A very limited time is needed to aminoglycosides also they are very operative to counter bacterial population. Such type of processes are recognized to a prime mechanism as inhibitors of protein synthesis, although other mode of action are associated for certain specific agents.
Mode of aminoglycoside resistance
There are several mechanisms of aminoglycoside resistance used by bacteria which are as follow. This mechanism can be chromosomally or plasmid mediated.
Reduced transport: This is one of the characteristic mechanism, limited transport via cell membrane (non plasmid mediated resistance) decreased transport can be tempted by means of coverage to the sub lethal concentrations of such antibiotics one of the examples include streptomycin resistance.
Reduced ribosomal binding: This is another important but not commonly used mechanism in which there is a limited access to the ribosomal subunit to avoid mistranslation of the protein and hence saving the bacteria. This mechanism may not be clinically important form of resistance.
Enzymatic modification: This is one of the important modes of resistance against aminoglycosides. It can be either chromosomally or plasmid mediated. The enzymes are present both in gram-negative and gram-positive bacteria. Basically three important types of enzymes are included comprising some subclasses: Adenylating enzymes (nucleotidyltransferases), phosphorylating enzymes (phosphotransferases) and acetylating enzymes (acetyltransferases). The liability of every aminoglycoside to particular enzymatic attack differs amongst every subclass. Even though cross-resistance is communal, there are alterations in the patterns of susceptibility. The drugs are stabilized by chemical alteration that reduces liability toward enzymatic damage. For example, chemically altered kanamycin produces amikacin that is further more resistant to the enzymatic hydrolysis. Such modifications are controlled by specific genes [3].
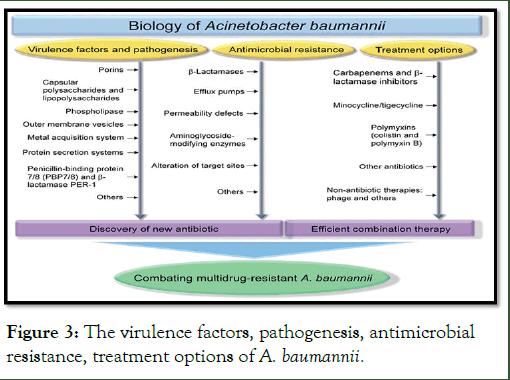
Figure 3: The virulence factors, pathogenesis, antimicrobial resistance, treatment options of A. baumannii.
Aminoglycoside resistance genes
There are basically three aminoglycoside-modifying enzymes all of these have been present in the Acinetobacter specie. The enzymes are O-phosphotransferases (APH) and Onucleotidyltransferases (ANT) and, that catalyse, respectively, phosphorylation and nucleotidylation (adenylation) of hydroxyl groups, and the third one is N-Acetyltransferases (AAC), thatleads to or catalyse acetylation of amino groups, hence making antibiotic inactive. Acinetobacter may contain several aminoglycoside resistance genes.
The genes for these enzymes are frequently present on the mobile elements, loke plasmids and transposons, and are shifted among the population of A. baumannii. The synthesis of AAC(3)-I, APH(3’)-VI and ANT(3’’)-I are present and found to as playing major role by several investigations on isolates of A. baumannii, but there are significant regional modifications in their genotypes.
Resistance for the aminoglycosides very frequently come across by aminoglycoside modifying enzymes, taking in count phosphotransferases, nucleotidyltransferases and acetyltransferases. More recently, 16S rRNA methylases, RmtD, RmtB, ArmA, Rmt andRmtA, have been stated amongst Enterobacteriaceae, Acinetobacter and Pseudomonas speci. There are some important genes that play important role in generating resistance against antibiotics. Such as aph(3’)-Ia, aph(3’)-Via, aac(3,)-Ia, aac(3’)-IIa, ant(2’)-Ia or aadB, ant(3’)-Ia, armA, aadA, aacC1 etc. these genes play important role in resistance operations. There are some other genes as well.
“aacC1”, “aadB” and “aph3” gene play a vital role in aminoglycoside resistance in A. baumannii. The product of “aacC1” gene is gene product is “Acetyletransferases”. This gene basically targets gentamicin. Product of “aadB” gene is “2’- Aminoglycoside nucleotidyltransferase”. Target of this gene are kanamycin, tobramycin and gentamicin as well, whereas the product of aph3 gene is "aminoglycoside 3'-phosphotransferase". Gene target is kanamycin and gentamicin.
Basic focus of this research is to check prevelance and correlation of aacC1, aadB and aph3 gene for aminoglycosides resistance in A. baumannii. This study would be informative for relationship between gene and resistance against aminoglycosides (specifically kanamicin and gentamicin) and in A. baumannii [4].
Rationale
Acinetobacter infections have developed as most important cause of hospital-acquired infections in the world. One of the most clinically substantial pathogens is A. baumannii because of the resistance developing speed to the current antimicrobial drugs and the capability of various A. baumannii strains to be survive on the hospital equipment and facilities for weeks. Extensive variety of infections are caused by Acinetobacter include respiratory tract infections, bacteremia, urinary tract infections, skin and soft tissue infections, osteomyelitis, and intracranial infections. Due to the limited therapeutic options for multidrug-resistant Acinetobacter infections the mortality related with A. baumannii infection in the Intensive Care Unit (ICU) setting is increasing. To develop novel therapeutics agents active against multidrug resistant strains is need of time. This study focus on the determination of aminoglycosides resistance pattern and the prevalence of aminoglycoside resistance genes in local clinical isolates of A. baumannii. The information obtained by this study would be utilized for designing the new drug analogues [5].
Objectives
Objectives of the study were to:
- Determine the antibiotic susceptibility of A. baumannii strains against aminoglycosides.
- Determine the prevalence of aminoglycoside resistance genes, and in A. baumannii local clinical isolates by PCR.
Literature Review
A study was conducted based on aminoglycoside resistance. The study included all clinical samples from Intensive Care Units (ICUs) patients with suspected hospital acquired infections. Positive cultures were identified by gram stain and complete biochemical identifications. Antibiotic susceptibility was carried out by disc diffusion method. Minimum Inhibitory Concentration determination (MIC) to amikacin and gentamicin was performed by microdilution method. Polymerase Chain Reaction (PCR) was performed for detection of aadB, aadA1, aphA6, aadC1 genes. Results stated that the study included 1,200 bacterial isolates from patients admitted to ICUs during the period of the study. A. baumannii represented 100 isolates from gram negative bacilli. The study of MIC of A. baumannii to amikacin and gentamicin revealed that 50 (50%) of the isolates had resistance by MIC study with 36 (72%) of those having High Level Aminoglycoside Resistance (HLAR) and 14 (28%) had non-high-level aminoglycoside resistance. The most common prevalent resistant genes among A. baumannii resistance to aminoglycosides was aadB (42%), followed by aphA6 (26%). Less prevalent genes were aadA1 (18%) and aacC1 (12%). There were 24 isolates with negative PCR for the studied genes. In a comparison between the prevalence of resistant genes among A. baumannii with HLAR and non HLAR, there was a non-significant increase of aphA6 (30.6%) and aadB (44.4%) in HLAR isolates compared to non HLAR (14.3%, 35.7%, respectively; p=0.2), with a significant increase in aacC1 28.6% in HLAR compared to 5.6% in non HLAR (p=0.02). comes to the conclusion that half of the clinical isolates of A. baumannii have resistance to the aminoglycosides amikacin and gentamicin. Most isolates have a high resistance level to aminoglycoside [6].
A study was conducted on aminoglycosides resistance, the consequences explained an ultimate association amongst presence of armA as well as AME encoding genes and resistance of A. baumannii against aminoglycosides. In development of Aminoglycoside resistance, it had fractional role along with the up-regulation of AbeM and AdeABC systems.
In another study the involvement of AdeIJK, AdeFGH and AdeABC Resistance-Nodulation-cell Division (RND)-type efflux systems to managed and the intrinsic resistance, it was built, from an entirely sequenced susceptible A. baumannii strain, a group of the isogenic mutant’s overexpression of every system resulting in introducing a point mutation in cognate regulator or a removal for pump by the replacement of allels. AdeFGH and AdeABC are strongly regulated and play role in acquirement of the antibiotic resistance as result of overproduction. It was revealed by Pairwise contrast of each derivative to parental strain. The AdeABC had a wide substrate range, along with fluoroquinolones, beta-lactams, macrolides, chloramphenicol, tetracyclines-tigecycline, and convened clinical resistance against the aminoglycosides. Considerably, when joined with enzymatic resistance to aminoglycosides, and carbapenems this pump further added in the synergistic manner of resistance level to the host. The AdeIJK was expressed innately and reason for essential resistance to same foremost drug classes as the AdeABC also fusidic acid and antifolates. Unexpectedly, the overproduction of AdeIJK plus AdeABC transformed structure of bacterial membrane, causing the reduced biofilm formation but not the motility. Plasmid transfer and natural transformation were reduced in the recipients overproducing AdeABC. Therefore, it looks as if that modification in expression of the efflux systems results into manifold alterations in linkage between the host and environment, addition to resistance against antibiotics [7].
Another study was conducted which about 4680 isolates taken from 45 hospitals were tested aimed at the exposure with antimicrobials, consuming the method of reference broth microdilution. Chosen separates or isolates of, Aminoglycoside-Modifying Enzymes (AMEs), 16S rRNA methyltransferases and carbapenemases were screened for genes encoding. Study comes to conclusion, Plazomicin presented activity against isolates of Enterobacteriaceae, counting the isolates having AMEs and a high proportion of the carbapenem-non-susceptible isolates. The Plazomicin also presented action against staphylococci.
In another study was made to explain epidemiological and genetic characteristics of strains obtained from a medical location in the Nepal. About 246 Acinetobacter specie isolates took from various patients, and were partitioned for MDR (Multi Drug Resistance). The multilocus sequence forms were assembled that leads to recognition of resistant genes. From 246 isolates of Acinetobacter specie 122 (49.6%) were multi-drug resistant A. baumannii, most of this was being resistant against the aminoglycosides, fluoroquinolones and the carbapenems except tigecycline and colistin. The following isolates give refuge to the blaOXA-23 blaNDM-1, and blaOXA-58. As well as 16S rRNA methylase gene armA. Multi-drug resistant A. baumannii isolates suitable for the Clonal Complex (CC1 and CC2) along with a unique clonal complex (CC149) that have shaken out all over in medical scenery in the Nepal. Genes encrypting or coding for a 16S rRNA methylase (ArmA) and carbapenemases (OXA and NDM-1) were harbored by the MDR.
In another study, Vulnerability or liability to the antimicrobials and biocides was tested in 49 isolates of A. baumannii that were clonally separate. Biological charge, in the positions of the nasty generation time, was recognized by the spectrophotometry technique. To obtain the comparative expression of the genes coding numerous porins and the efflux pumps, RT–PCR was performed. It was determined that decreased liability to the biocides is linked to co-resistance against the ciprofloxacin, aminoglycosides, tetracycline and carbapenems. There is a biological cost for acquirement of decreased susceptibility for the benzalkonium chloride. Comparative expression of the genes associated to certain or porins (reduced expression) or efflux pump genes (increased expression) get effected by exposure of the biocides.
A study was made to assess assortment of the antibiotic resistance genes, for which an aggregate of the 160 CRAB strains were separated at the tertiary care hospitals of Pakistan which primarily had blaOXA-23 gene. This study concluded that despite the CRAB clones containing blaOXA-23 have been reported previously in Pakistani hospitals, screening of the genetic determinants accountable for other antimicrobial agents is significant for emerging an effective investigation and facilitation system for the infection management [8].
Methodology
Bacterial strains characterization
Study location: Total 50 bacterial strains were collected from the Mayo Hospital, services institute of medical sciences and children hospital and the institute of child health. Characterization of clinical isolates was made by checking their characteristic growth pattern of particular media, oxidase test, gram staining, biochemical test, API and antibiotic susceptibility according to CLSI guidelines. Extraction of DNA was done department of microbiology of molecular biology, university of the Punjab. The PCR and Gel electrophoresis was done at department of biochemistry, Kinnaird College for Women.
Isolation of A. baumannii: Several types of samples were used to isolate bacterial strains such as blood, thoracic swab, ear swab, and pleural fluids, CSF and urine. Swabs and CSF samples were cultured on blood agar and MacConkey agar on same day. Urine samples were cultured on CLED agar after 24 hours of incubation. Whereas the blood samples were cultured on Blood and MacConkey agar after. All the samples after streaking were placed on overnight incubation at 36°C. Once the growth was obtained the next step proceeds as follow.
Characterization of A. baumannii
Gram staining: Gram staining was performed according to standard gram staining protocol. In the very first normal saline was applied on the slides. An isolated colony of the bacteria was picked up and then applied to saline and mixed well. The next step of it was to place the slides into oven for proper drying. This drying leaded to bacterial colony fixation on slide. After fixation the slide was covered with crystal violet stain for one minute at room temperature, risen with water after one minute. Crystal violet stained the bacterial colony or specimen. The next step was to apply gram iodine for one minute to fix the stain. Risen with water after one minute. In the next step acid alcohol was applied for 15 seconds, as the gram positives have low permeability, did not let out stain easily as compared to gram negatives who have high permeability. That’s why the gram negative gets stained pink because of high permeability of their outer membrane the stain went out of specimen. After wash next step was to apply Safranin for 2 minutes and risen properly with water. Hence the plates were ready to visualize under microscope. Gram positive stained purple and gram-negative stained pink. Whereas the Acenitobactor stained as gram negative.
Oxidase test: For gram negative we perform “Oxidase test” according to standard procedure. The purpose of “Oxidase” was a quick identification of bacterial cytochrome oxidase. It was provided ready to use. It was composed of a 0 .3% N, N, Nl, Nltetramethyl- p-phenylenediamine in a 0.003 M buffer comprising organic stabilizers and reducing agents. Aerobic bacteria have Electron transport system in it as in mitochondria. The ETS of aerobic bacteria have Cytochrome C Oxidase enzyme. TMPD (Tetramethyl-p-phenylenediamine) transfer an electron to cytochrome C Oxidase as a result there was a transformation into TMPD radical producing purple color whereas this reaction does not take place in anaerobic ones that’s why producing no color. A. baumannii gives negative oxidase [9].
Biochemical identification: After oxidase we perform biochemical test for colonies on “Macckonkey agar plates” for confirmation of Acinetobacter. We performed four identification tests like Citrate, Urease, TSI (triple sugar iron test) and MIO/SIM media test. An isolated colony of oxidase negative bacteria was picked with loop and firstly applied to citrate agar, urease agar base, TSI agar an in zigzag manner and vertically straight in MIO/SIM medium respectively. These were placed for 24 hours of incubation to obtain results. Citrate has Bromothymol blue in it that cause change in color if there were positive and remain green if results got negative. Acinetobacter gave citrate positive. The Urease has phenol red in it and it gave pink color at positive and yellow for the negative. This test helped to differentiate Acinetobacter from other gram negatives such as Klebsiella, citro and protease etc. TSI (Triple Sugar Iron) test was made to confirm the Acinetobacter. If the bacteria were lactose fermenting it gave cause the pH acidic and turned its color to yellow, if bacteria were fermenting lactose but, in less amount, it results in both acidic and alkaline in pH caused red yellow combination. If the bacteria were non-fermenting it gave result in alkaline PH with red color. Only yellow ones could be E. coli, Klebsialla and enterococci, whereas bacteria that caused red yellow combination could be citro, Salmonella and serratia. On the other hand, only red ones could be pseudomonas or Acinetobacter. Final step of biochemical test was MIO/SIM medium to identify motility of organism.
API (Analytical Profile Test): A very fine method for identification of micro-organism on their species level was use of API test kits which used to identify the gram positive, gram negative bacteria and yeast according to standard protocol. This system suggests a wide and robust database that is available by Internet-based “API WEB” service. Several tests were performed at the same time in the API 10S kit Such as, LCD, ARA, ONPG, CIT, 6ODC, H2S, URE, ARA, ONPG, CIT. Whereas. We could get their results right after incubation. In case of following tests different reagents were applied after incubation like GLU, NO2, H2S, TDA and IND to obtain results.
To perform API first of all, took normal saline, picked an isolated colony and mixed well in the normal saline in tube. After homogeneous shaking the solution mixture was applied to API10S kit. For CIT test both of the cupule and tube were filled. For other tests, only tubes were filled. For the test LDC, ODC, H2S and URE created anaerobiosis by application of mineral oil. Also, the bacterial suspension was cultured on MacConkey plate as positive control and place it for incubation for 24 hours. As incubation period was completed put out kit and MacConkey agar plates from incubator and checked the growth on sub cultured media also start Interpretation of test Kit. For interpretation there were following tests that was required to do as:
- TDA test: One drop of TDA reagent was applied to cupule. Reddish brown color indicates positive result for mark sheet.
- IND test: One drop of James reagent was applied. Positive ones indicate pink color to be recorded on result sheet.
- NO2 test: One drop of NIT1 and NIT2 reagent in GLU, the red color indicates positive result to be marked on test sheets.
API test specifically for colonies on “blood agar plates”.
Drug susceptibility testing: After conformation of organism, a range of antibiotics was applied to check antibiotic resistance and susceptibility to select multi drug resistant A. baumannii. Swabing of bacterial colony is made on MH-media plates. Punches of antimicrobial drugs were applied to plates after swabbing and placed in incubator at 36°C overnight. Following punches of drugs were applied on plates and read according to CLSI guidelines (Table 2).
| Antibiotics | Symbol | Concentration | Resistant | Intermediate | Susceptible |
|---|---|---|---|---|---|
| Colistin sulphate | COL | 25 µg | ≤ 11 | 45639 | ≥ 14 |
| Polymyxin | POL | 30 µg | ≤ 11 | 45576 | ≥ 12 |
| Sulbactam/Cefoperazone | CPS | 10/5 µg | ≤ 17 | 18-20 | ≥ 21 |
| Augmentin | AUG | 20/10 µg | ≤ 13 | 14-17 | ≥ 18 |
| Cefotaxime | CTX | 30 µg | ≤ 14 | 15-22 | ≥ 23 |
| Meropenem | MER | 10 µg | ≤ 14 | 15-17 | ≥ 18 |
| Tazobactam | TZP | 100/10 µg | ≤ 17 | 18-20 | ≥ 21 |
| Ceftriaxone | CTR | 30 µg | ≤ 13 | 14-20 | ≥ 21 |
| Ceftazidime | CAZ | 30 µg | ≤ 14 | 15-17 | ≥ 18 |
| Ciprofloxacin | CFP/CPR | 5 µg | ≤ 15 | 16-20 | ≥ 21 |
| Clathromycin | CLM | 15 µg | ≤ 13 | 14-17 | ≥ 18 |
| Trimethoprim-sulfamethoxazole | COT | 1.25/23.75 µg | ≤ 10 | 45611 | ≥ 16 |
| Cefuroxime | CXM | 30 µg | ≤ 14 | 15-22 | ≥ 23 |
| Moxifloxacin | MXF | 5 µg | ≤- | - | ≥ 18 |
| Amikacin | AMK | 30 µg | ≤ 14 | 15-16 | ≥ 17 |
| Ciprofloxacin | CIP | 5 µg | ≤ 32 | 33-34 | ≥ 35 |
| Doxycyline | DO | 30 µg | ≤ 9 | 45577 | ≥ 13 |
| Ceftriaxone | CRO | 30 µg | ≤ 13 | 14-20 | ≥ 21 |
Table 2: Antibiotic sensitivity range according to CLSI guidelines.
Isolation of genomic DNA
Isolation of the genomic DNA of A. baumannii from screened bacterial strain was done by using standard phenol/chloroform method. There are several steps of genomic DNA purification protocol. For extraction of gram negative DNA, 1 ml of the cell suspension was centrifuged for 2 minutes at 8000 g used to pellet cells. In the next step the removal of the supernatant was placed, cells were washed along with 400 μl of STE buffer (100 mM Nacl, 1 mM EDTA, 10 mM tris-Hcl,PH8.0). Re-suspension of pellets was performed in 200 μl TE buffer (1 mM EDTA, 10 mMTris/HCl, PH8.0). Formerly 100 μl of Tris-saturated phenol (pH 8.0) was added in tubes, leaded by step of vortex-mixing of 60 sec for bacteria. After that Samples were centrifuged for 5 min at 13000 g at 4°C for separation between organic and aqueous phase. 160 μl of upper aqueous phase was shifted to a new 1.5 ml tube. Addition of 40 μl TE buffer was placed to make 200 μl along with 100 μl chloroform and centrifuged at 13000 g for 5 min at 4°C. Purification of the lysate was made by chloroform extraction till white interface no longer exist, this process was repeated two to three times. 160 μl of the upper aqueous phase was shifted to clean 1.5 ml tube. 40 μl TE and 5 μl RNase (at 10 mg/ml) were added and then incubated at 37°C for 10 min for RNA digestion. Then 100 μl chloroform was added in tube, properly mixed and centrifuged at 13000 g for 5 min at 4°C. 150 μl of upper aqueous phase was shifted to clean 1.5 ml tube. Pure DNA was present in aqueous phase. The DNA concentration was checked by running the DNA on gel and stored at 20°C [10].
Agarose gel electrophoresis
1% Agarose gel was prepared to visualize the result of gene that was amplified. For this purpose, following solutions were used:
Gel loading dye: Commercially available xylene dye was used along with samples during loading.
TAE buffer: For 1 L 50X TAE buffer stock preparation, 57.1 ml of glacial acetic acid, 242 g of Tris base and 100 ml (0.5 M). EDTA (pH 8) were dissolved in the 1 L distilled water. 10 ml of 50X TAE Buffer was diluted with 490 ml distilled water to make 1xTAE buffer working solution.
Ethidium bromide: The available commercially prepared ethidium bromide having 0.5 μg/ml concentration was used.
Gel electrophoresis protocol: The concentration of Isolated DNA was checked on 1% agarose gel. 1 g agarose was added in 100 μl of 1xTAE buffer and heated to dissolve proper. After a bit lowering in temperature of gel commercially available 8 μl ethidium bromide was added into the gel and poured into casting tray. Once the gel solidified, it was placed in 1xTAE buffer tank, removed the comb and aaded more 1x TAE buffer so the gel got properly covered by buffer and loaded the DNA samples along with gel loading dye (xylene). Gel was run at 80 volt for 35 minutes. Then gel was takene out of the tank and placed in Uitraviolet trans-illuminator (Universal Hood II BIORAD) to visualize bands.
Primer designing
Gene aacC1 (AC No. KR610434) with 490 base pairs, Gene aadB (AC No. HQ148722) with 500 base pairs and gene aph3 (AC No. KY123748) with 489 base pair were selected to be amplified for detection of aminoglycosides resistance in Acenitobacter. Fasta sequence of these genes was retrieved from NCBI. Primers were designed for the selected genes by using Primer 3 and their properties were calculated by Oligo Calc (Table 3) [11].
| S. no. | Genes | Primers | Length of column | TM (°C) | GC % |
|---|---|---|---|---|---|
| 1 | aacC1 | FP:5’-GTGGCTCAAGTATGGGCATCATT-3’ | 23 | 58 | 48 |
| RP:5’-AAT TGTTAG GTGGCG GTACTTGG-3’ | 23 | 58 | 48 | ||
| 2 | aadB | FP: 5’-CGATGTTACG CAGCAGCAACG-3’ | 21 | 60 | 57 |
| RP: 5’-TGGACGAATTGTTAGGCCGCATA-3’ | 23 | 58 | 48 | ||
| 3 | aph3 | FP: 5’-TCGAAAAATACCGCTGCGTAAAAGATACG-3’ | 29 | 61 | 41 |
| RP:5’-TCTTTAAAAAATCATACAGCTCGCGCGG-3’ | 28 | 61 | 43 |
Table 3: Sequence and propertied of primers of “aacC1”, “aadB” and “aph3” genes.
Primer dilution
Initially the primers were in lyophilized state. A stock solution was prepared from lyophilized state. A short spin was given to primers to remove pallet from cap. To determine the amount of water added into the lyophilized state we found nmole of primers mentioned on main tube and multiplied it with 10. That was exact amount of deionized, distilled or injection water added for 100 μl stock solution. For aacC1 gene forward primers 21.88 mole was mentioned on tube so it get 219 μl of injection water and for reverse primer was of 21.65 mole so it got 217 μl injection water. Same as primer for aadB gene forward primer 23.65 mole and reverse primer 21.38 mole so, 237 μl and 214 μl injection water was added to make stock solution. For aph3 forward primer 16.08 moles and reverse primer 17.71 moles, volume of diluent was 161 μl and 177 μl respectively. The working solution was made by 10 μl of stock solution and 90 μl of injection water. This working solution was ready to run PCR (polymerase chain reaction).
PCR amplification: The PCR amplification of the genes (multiplex PCR amplification) was done using standard protocols. PCR reaction was carried out by setting conditions of denaturation, annealing and extension. Polymerase chain reaction based on replication of DNA strand by elongation of the complementary strand started from one pair of thoroughly space out chemically manufactured oligonucleotide primers.
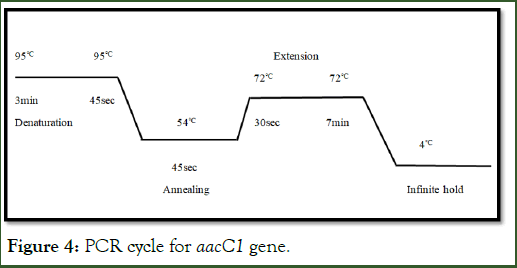
aacC1 gene amplification: Basic technique of the PCR comprises repetitive cycles of amplifying particular sequence of nucleic acid. A cycle of PCR consists of three steps: (1) denaturation of DNA at 95°C for 3 min-45 sec that results in separation of double stranded DNA, (2) primer annealing at relatively lower temperature at 54°C for 45 sec, (3) and finally extension at 72°C for 30 sec to 7 min and then leaves of to 4°C in which extension of sequence between primers takes place. In result of each cycle end the quantities of the PCR product was doubled. A thermal cycler is used to precede this procedure for 30 cycles. The annealing temperature should be 3-5°C below then Tm (melting temperature). During this quantity of total reaction was 10 μl. The components of this reaction mixture are as follow (Table 4).
| S. no. | Components of reaction | Quantity |
| 1 | Water | 5 µl |
| 2 | Master mix (DNTPs, buffer, Mgcl2) | 1.75 µl |
| 3 | Forward primer | 1 µl |
| 4 | Reverse primer | 1 µl |
| 5 | Template | 1 µl |
| 6 | Taq poly | .25 µl |
| Total | 10 µl |
Table 4: components of PCR reaction for “aacC1”, “aadB” and “aph3” gene.
Above mentioned components in Table 4 were used to run standard PCR protocol. As mentioned in Figure 4 at initial stage in the step of denaturation, temperature was 95°C for 3 min followed by 30 cycles at following conditions, denaturation for 45 sec. After that annealing at 540 for 45 sec and finally extension at 72°C from 30 sec to 7 min. After extension the temperature is lowered till 4°C on infinite hold. PCR products are transported to Freezer or directly could be applied to gel [12].
Figure 4: PCR cycle for aacC1 gene.
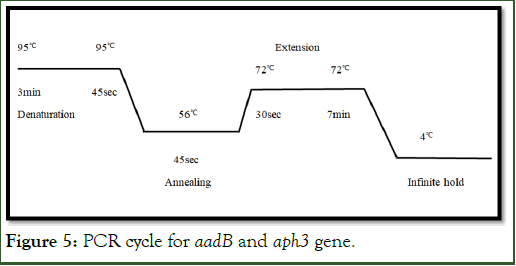
aadB and Aph3 gene amplification: For amplification of aph3 gene in A. baumannii DNA, the conditions were optimized in PCR. Three basic steps were involved to target aph3 gene as shown n Figure 5 (1) Denaturation of DNA at 950 from 3 min and then 45 sec, (2) Annealing at 560 for 45 sec that leads to primer binding at specific target helps in amplification and (3) Final step extension at 720 for 30 sec and headed to 7 min for final extension. As a result, multiple copies of amplified target gene were obtained with size of 489 bps. A total of 30 cycles were involved with 10 ul volume in PCR reaction.
Figure 5: PCR cycle for aadB and aph3 gene.
Agarose gel electrophoresis: 1% gel was prepared by method described above in section 3.4. For visualization of amplified bands of respective genes in clinical isolates of A. baumannii, the PCR products were loaded on gel along with xylene dye and ran at 100 volts for 50 minutes with 100 bps ladder. After specific time gel was removed and placed in ultraviolet trans-illuminator (Universal Hood II BIO-RAD) to visualize bands [13].
Results
Growth pattern of A. baumannii
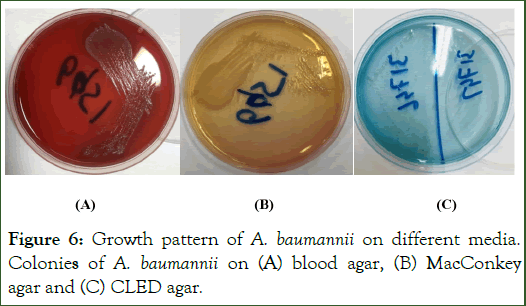
A. baumannii was isolated from different samples i.e. blood samples, thoracic swabs, ear swabs, pleural fluid, CSF and Urine samples. A. baumannii showed light pink color, normal sized, less mucoid and well isolated colonies on MacConkey agar as shown in Figure 6. It also showed growth on Blood agar as well as on CLED agar specifically for microbes present in urine sample. On CLED agar, A. baumannii showed opaque, colourless, smooth colonies while on blood agar, it showed non hemolytic, opaque, circular grey colored colonies as showed in Figure 6.
Figure 6: Growth pattern of A. baumannii on different media. Colonies of A. baumannii on (A) blood agar, (B) MacConkey agar and (C) CLED agar.
Gram staining
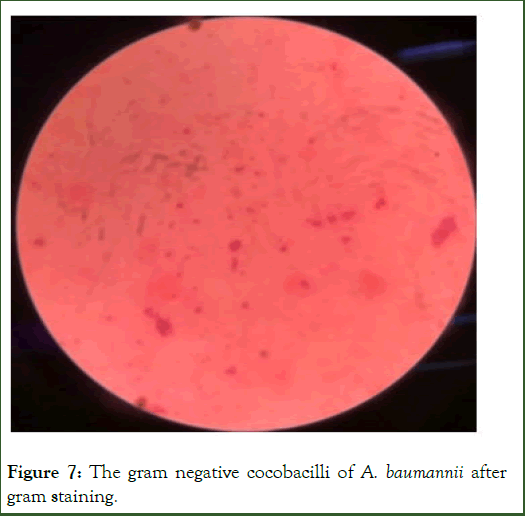
After isolation of A. baumannii, gram staining was performed. It showed pink colored cells under microscope characteristic of gram negative bacteria as shown in Figure 7. The morpholog of the A. baumannii showed under microscope was coccobacilli.
Figure 7: The gram negative cocobacilli of A. baumannii after gram staining.
Oxidase test
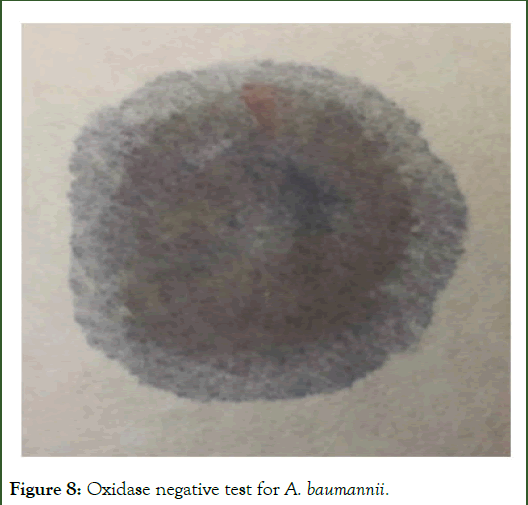
Oxidase test is used to detect the presence of cytochrome oxidase enzyme. A positive result by is depicted by the appearance of purple colour while colourless appearance suggest the negative result. A. baumannii showed the colourless appearance showing its oxidase negative status as shown in Figure 8.
Figure 8: Oxidase negative test for A. baumannii.
Biochemical test
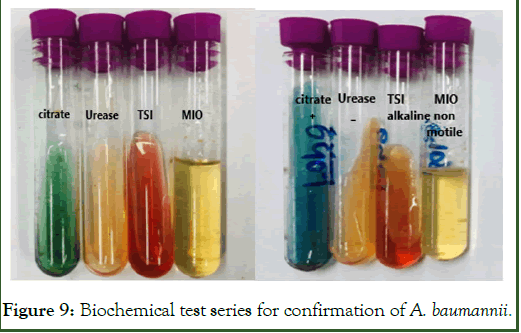
During characterization of bacterial colonies different other biochemical tests were performed. Citrate test gave positive result turning blue, urease negative with no change in colour, TSI test gave alkaline slant in red and non-motile in result of MIO test, gave confirmation that the bacterial strain is Acinitobactor (Figure 9).
Figure 9: Biochemical test series for confirmation of A. baumannii.
API
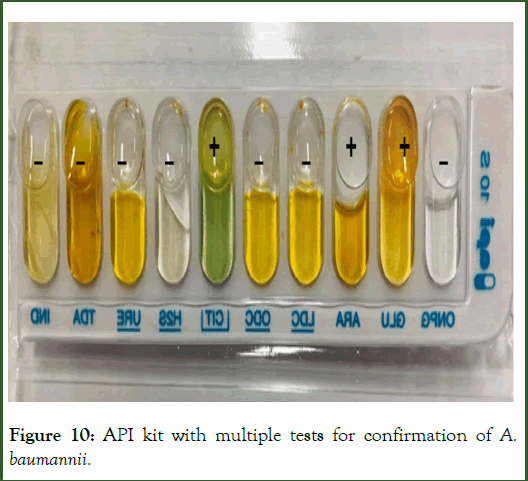
API 10S kit was used for the final characterization of the A. baumannii. API kit is able to perform 10 types of different test required for characterization simultaneously. The isolates were ONPG negative, GLU positive, ARA positive, LDC negative, ODC negative, CIT positive, H2S negative, URE negative, TDA negative and IND negative. The results were entered in API Software, where software gives confirmation that the bacterial isolate was of A. baumannii. Also, A. baumannii reference strain was incubated for 24 hours as positive control. The result of API kit is shown in Figure 10.
Figure 10: API kit with multiple tests for confirmation of A. baumannii.
Antimicrobial susceptibility of A. baumannii
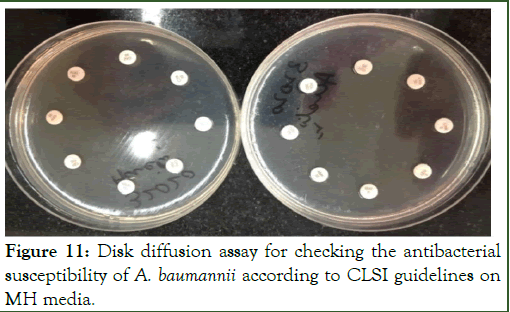
Antibiotic susceptibility of A. baumannii against the antimicrobial drugs shown in Table 5 was checked on MH-media was performed. After swabbing of bacterial strains, antimicrobial disks were applied and incubated at 37°C for 24 hours and results were then recorded according to the CLSI guidelines as shown in Figure 11.
Figure 11: Disk diffusion assay for checking the antibacterial susceptibility of A. baumannii according to CLSI guidelines on MH media.
Multiple categories of antimicrobial agents were used to check the sensitivity pattern for A. baumannii strains. Different drugs belonging to lipopeptide, aminoglycoside, beta-lactum, cephalosporins, carbapenems, macrolid, sulphonamide, floroquinolones and tetracyclins were used as mentioned below in Table 5. The results of antimicrobial sensitivity testing of all the 50 A. baumannii is given in Table 5.
During sensitivity reading, multiple frequencies of resistance were obtained by different drugs as shown in below Table 3. A. baumannii strains showed 100% resistance against gentamicin, ceftriaxone,and doxycyclin antibiotics. The second highest resistance frequency was 94% against cefotaxime. Third highest resistance frequency was 86% against sub-lactum and cefuroxime. A. baumannii strains showed 84% and 80% resistance against Amikacin and Meropenem, respectively. Middle range resistance frequency were 76%, 74%, 66%, 60%, 53%, 46%, 40% against tazobactum and augmentin, clarithromycin, cefipime, trimethoprime-sulfamethoxazole, moxifloxacin and levofloxacin respectively. The lowest resistance frequency of 34% was obtained against ciprofloxacin. All the A. baumannii strains were sensitive to colistin and polymyxin [14].
| Antimicrobial category | Antibiotics | Symbol | Resistant/total 50 strains | Resistance frequency (%) |
|---|---|---|---|---|
| Lipopeptide | Colistin | COL | 0 | 0 |
| Polymyxin | POL | 0 | 0 | |
| Aminoglycosides | Amikacin | AMK | 38 | 0.76 |
| Gentamiin | CN | 38 | 0.76 | |
| Kanamycin | KAN | 35 | 0.7 | |
| Beta-lactum | Sub lactum | CPS | 7 | 0.14 |
| Tazobactum | TZP | 38 | 0.76 | |
| Augmentin | AUG | 37 | 0.74 | |
| Cephalosporin | Cefotaxime | CTX | 47 | 0.94 |
| Ceftriaxone | CRO/CTR | 50 | 1 | |
| Cefipime | CFP | 30 | 0.6 | |
| Cefuroxime | CXM/CRX | 43 | 0.86 | |
| Carbapenem | Meropenem | MEM | 40 | 0.8 |
| Macrolid | Clarithromycin | CLM | 33 | 0.66 |
| Sulfonamides | Trimethoprime-sulfamethoxazole | COT | 26 | 0.53 |
| Fluroquinolones | Ciproflaxacin | CIP | 17 | 0.34 |
| Moxifloxacin | MXF | 23 | 0.46 | |
| Levofloxacin | LEV | 20 | 0.4 | |
| Tetracycline | Doxycyclin | DO | 50 | 1 |
Table 5: % frequencies of resistance against different antimicrobial drugs in A. baumannii strains.
Multiple drug resistance pattern of A. baumannii: Most of the A. baumannii strains contain resistance to more than one antibiotic. The Table 6 gives information about pattern of multiple drug resistance pattern by the isolated A. baumannii strains.
| Combination of drugs classes | Number of resistant strains | (%) Frequency |
|---|---|---|
| Aminoglycosides, beta-lactam, cephalosporins, carbapenem, sulfonamide | 1 | 2 |
| Aminoglycosides, beta-lactam, cephalosporins, carbapenem, macrolid | 1 | 2 |
| Aminoglycosides, cephalosporins, carbapenem, macrolid, sulfonamide | 1 | 2 |
| Aminoglycosides, cephalosporins, carbapenem, tetracyclin | 6 | 12 |
| Aminoglycosides, beta-lactam,cephalosporins, carbapenem, tetracyclin | 5 | 10 |
| Aminoglycosides, cephalosporins, carbapenem, fluroquinolones, tetracyclin | 4 | 8 |
| Aminoglycosisdes, beta-lactam, cephalosporins, carbapenem, fluroquinolones, tetracyclin | 13 | 26 |
| Aminoglycosides, fluroquinolones, tetracycline | 2 | 4 |
| Aminoglycosides | 3 | 6 |
| Cephalosporins, sulfonamide | 3 | 6 |
| Beta-lactam, cephalosprins, carbapenem, fluroquinolones, sulfonamide | 6 | 12 |
| Cephalosprins | 1 | 2 |
| Beta-lactam | 2 | 4 |
| Cephalosporins, carbapenem, fluroquinolones, tetracycline | 2 | 4 |
| Total strains | 50 | 100 |
Table 6: Resistance pattern of A. baumannii against multiple drug.
Aminoglycosides susceptibility of A. baumannii: Out of total 50 drug resistance strains of A. baumannii, 38 (76%) showed resistance against aminoglycoside as shown in Table 5. Three different aminoglycoside drugs i.e. amikacin, gentamicin and kanamycin were used to check the sensitivity of A. baumannii [15].
Table 7 is representing the sensitivity and resistance against amikacin, gentamicin and kanamycin in A. baumannii strains (A1-A38). All the 38 strains (100%) were resistant to both amikacin and gentamicin while 3 strains out of 38 were sensitive to the Kanamycin as shown in Table 8
| A. baumannii strains | Amikacin (AK) | Gentamicin (GEN) | Kanamycin (KAN) |
|---|---|---|---|
| A1 | R | R | R |
| A2 | R | R | R |
| A3 | R | R | R |
| A4 | R | R | R |
| A5 | R | R | R |
| A6 | R | R | R |
| A7 | R | R | R |
| A8 | R | R | R |
| A9 | R | R | R |
| A10 | R | R | R |
| A11 | R | R | R |
| A12 | R | R | R |
| A13 | R | R | S |
| A14 | R | R | R |
| A15 | R | R | R |
| A16 | R | R | R |
| A17 | R | R | R |
| A18 | R | R | R |
| A19 | R | R | R |
| A20 | R | R | R |
| A21 | R | R | R |
| A22 | R | R | R |
| A23 | R | R | R |
| A24 | R | R | R |
| A25 | R | R | R |
| A26 | R | R | R |
| A27 | R | R | R |
| A28 | R | R | R |
| A29 | R | R | R |
| A30 | R | R | R |
| A31 | R | R | R |
| A32 | R | R | R |
| A33 | R | R | R |
| A34 | R | R | R |
| A35 | R | R | R |
| A36 | R | R | R |
| A37 | R | R | S |
| A38 | R | R | S |
Table 7: Aminoglycosides susceptibility in A. baumannii strains.
| Combined resistance | No. of Isolates | % frequency |
|---|---|---|
| AMK, GEN | 3 | 7 |
| AMK, GEN, KAN | 35 | 92 |
| Total | 38 | 100 |
Table 8: Resistance of aminoglycosides combination in A. baumannii.
Genomic DNA extraction
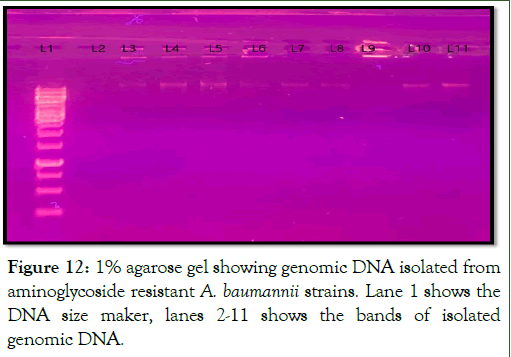
The genomic DNA of the aminoglycoside resistance strains were extracted and observed on 1% agarose gel under UV transilluminator. The bands of genomic DNA observed on gel is shown in Figure 12.
Figure 12: 1% agarose gel showing genomic DNA isolated from aminoglycoside resistant A. baumannii strains. Lane 1 shows the DNA size maker, lanes 2-11 shows the bands of isolated genomic DNA.
Detection of aminoglycoside resistance genes in A. baumannii
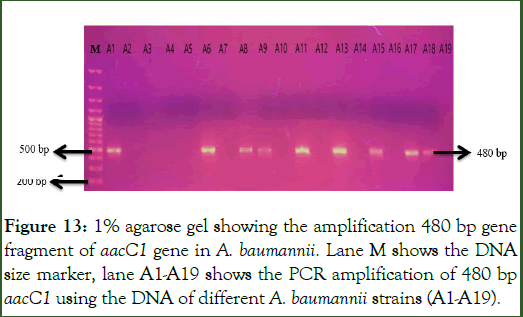
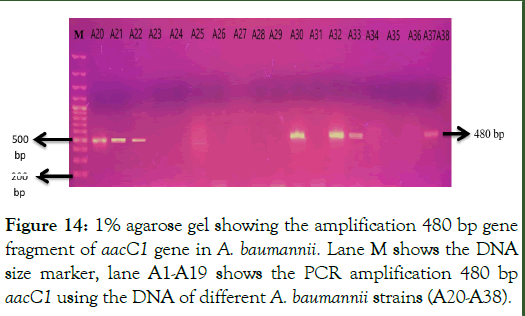
PCR amplification of aacC1 gene: The gene aacC1 which codes the Acetyleferases enzyme was amplified using the primers as shown in Table 3 in the total 38 aminoglycoside resistant A. baumannii strains. PCR products were loaded on 1% agarose gel for visualization of amplified gene fragment at specific size of 480 base pairs of aacC1 gene. Out of total 38 resistant strains, the 480 bp gene fragment of aacC1 gene was present in 16 out of 38 aminoglycoside resistant strains as shown in Figures 13 and 14.
Figure 13: 1% agarose gel showing the amplification 480 bp gene fragment of aacC1 gene in A. baumannii. Lane M shows the DNA size marker, lane A1-A19 shows the PCR amplification of 480 bp aacC1 using the DNA of different A. baumannii strains (A1-A19).
Figure 14: 1% agarose gel showing the amplification 480 bp gene fragment of aacC1 gene in A. baumannii. Lane M shows the DNA size marker, lane A1-A19 shows the PCR amplification 480 bp aacC1 using the DNA of different A. baumannii strains (A20-A38).
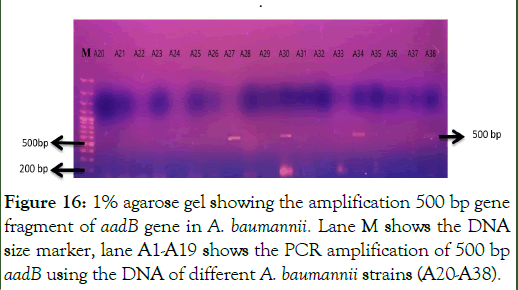
PCR amplification of aadB gene: The gene aadB that codes for “2’-Aminoglycosidenucleotidyltransferase” enzyme was amplified using the primers as shown in Table 3 in the total 38 aminoglycoside resistant A. baumannii strains. PCR products were loaded on 1% agarose gel for visualization of amplified gene fragment at specific size of 500 base pairs of aadB gene. Out of total 38 resistant strains, the gene 500 bp gene fragment of aadB gene was present in 13 strains as shown in Figures 15 and 16 [16].
Figure 15: 1% agarose gel showing the amplification 500 bp gene fragment of aadB gene in A. baumannii. Lane M shows the DNA size marker, lane A1-A19 shows the PCR amplification of 500 bp aadB using the DNA of different A. baumannii strains (A1-A19).
Figure 16: 1% agarose gel showing the amplification 500 bp gene fragment of aadB gene in A. baumannii. Lane M shows the DNA size marker, lane A1-A19 shows the PCR amplification of 500 bp aadB using the DNA of different A. baumannii strains (A20-A38).
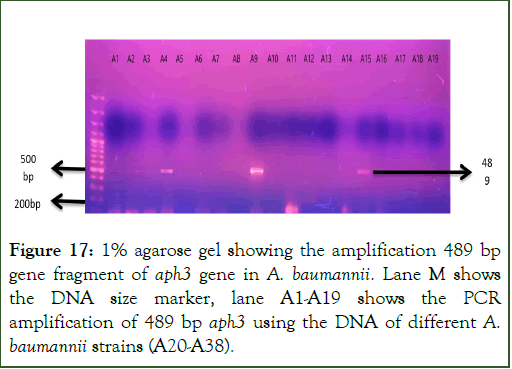
PCR amplification of aph3 gene: The gene aph3 which codes for “2’-Aminoglycosidenucleotidyltransferase” enzyme was amplified using the primers as shown in Table 3 in the total 38 aminoglycoside resistant A. baumannii strains. PCR products were loaded on 1% agarose gel for visualization of amplified gene fragment at specific size of 489 base pairs of aph3 gene. Out of total 38 strains, the gene 489 bp gene fragment of aph3 gene was present in 4 strains as shown in Figures 17 and 18 [17].
Figure 17: 1% agarose gel showing the amplification 489 bp gene fragment of aph3 gene in A. baumannii. Lane M shows the DNA size marker, lane A1-A19 shows the PCR amplification of 489 bp aph3 using the DNA of different A. baumannii strains (A20-A38).
Figure 18: 1% agarose gel showing the amplification 489 bp gene fragment of aph3 gene in A. baumannii. Lane M shows the DNA size marker, lane A1-A19 shows the PCR amplification of 489 bp aph3 using the DNA of different A. baumannii strains (A20-A38).
Following Table 9 represents % frequency of specific genes that are responsible for aminoglycosides resistance in A. baumannii strains. aacC1 gene is responsible for aminoglycoside resistance in 17 strains. Whereas the aadB gene is responsible for aminoglycoside resistance in 13 strains and aph3 in 4 strains.
| S. no. | Genes | Aminoglycoside modifying enzyme | Number of strains having gene | (%) |
| 1 | aacC1 | “Acetyletransferases” | 17 | 0.44 |
| 2 | aadB | “2’-Aminoglycosidenucleotidyltransferase” | 13 | 0.34 |
| 3 | Aph3 | “2’-Aminoglycosidenucleotidyltransferase” | 4 | 0.105 |
Table 9: Frequency of resistance genes among aminoglycosides resistant A. baumannii.
There were some A. baumannii strains in which more than one resistance gene was detected. Table 10 shows the pattern of resistance genes detected in aminoglycoside resistant A. baumannii strains (Table 11).
| Resistance gene detected | No. of A. baumannii strains | % frequency |
|---|---|---|
| aacC1 | 9 | 23 |
| aabB | 6 | 15 |
| aph3 | 2 | 5.3 |
| aacC1, aadB | 5 | 13 |
| aacC1, aph3 | 0 | 0 |
| aadB, aph3 | 0 | 0 |
| aacC1, aadB, aph3 | 2 | 5.2 |
| None of these genes detected | 14 | 36.8 |
| Total resistant strains | 38 |
Table 10: Responsible genes detected in aminoglycoside resistant A. baumannii strains.
| A. baumannii strains | Resistance to antibiotics | Resistance genes detected |
| A1 | AK, GEN, KAN | aacC1 |
| A2 | AK, GEN, KAN | aadB |
| A3 | AK, GEN, KAN | _ |
| A4 | AK, GEN, KAN | aph3 |
| A5 | AK, GEN, KAN | _ |
| A6 | AK, GEN, KAN | aacC1, aadB |
| A7 | AK, GEN, KAN | _ |
| A8 | AK, GEN, KAN | aacC1 |
| A9 | AK, GEN, KAN | aacC1, aadB, aph3 |
| A10 | AK, GEN, KAN | aadB |
| A11 | AK, GEN, KAN | aacC1, aadB |
| A12 | AK, GEN, KAN | aadB |
| A13 | AK, GEN | aacC1, aadB |
| A14 | AK, GEN, KAN | _ |
| A15 | AK, GEN, KAN | aacC1, aadB, aph3 |
| A16 | AK, GEN, KAN | _ |
| A17 | AK, GEN, KAN | _ |
| A18 | AK, GEN, KAN | aacC1, aadB |
| A19 | AK, GEN, KAN | _ |
| A20 | AK, GEN, KAN | aacC1 |
| A21 | AK, GEN, KAN | aacC1 |
| A22 | AK, GEN, KAN | aacC1 |
| A23 | AK, GEN, KAN | _ |
| A24 | AK, GEN, KAN | _ |
| A25 | AK, GEN, KAN | aacC1 |
| A26 | AK, GEN, KAN | _ |
| A27 | AK, GEN, KAN | aadB |
| A28 | AK, GEN, KAN | _ |
| A29 | AK, GEN, KAN | aph3 |
| A30 | AK, GEN, KAN | aacC1, aadB |
| A31 | AK, GEN, KAN | _ |
| A32 | AK, GEN, KAN | aacC1 |
| A33 | AK, GEN, KAN | aacC1 |
| A34 | AK, GEN, KAN | aadB |
| A35 | AK, GEN, KAN | _ |
| A36 | AK, GEN, KAN | _ |
| A37 | AK, GEN | aacC1 |
| A38 | AK, GEN | aadB |
Table 11: Pattern of antibiotic resistance and responsible genes detected among aminoglycoside resistant A. baumannii strains.
Discussion
A. baumannii is Multidrug-Resistant (MDR) pathogen that is responsible for nosocomial infections, specifically in the Intensive Care Units (ICUs) and patients with immune compromised situation and central venous catheters. A. baumannii has developed antimicrobial resistance for broad spectrum that is connected with developed mortality rate amongst the infected patients as compared to other. The clinical impact of the resistant strains are related with increases in both mortality and length of stay in hospital. A. baumannii can cause a range of infections; mostly involved respiratory tract, specifically ventilator-associated pneumonia, A. baumannii has developed several antibiotic resistance mechanisms, with growing harmful effects of its pathogenic potential and expressing a vital trial for the patients and healthcare providers [18].
Efficiency of cure depends mostly on exact antimicrobial combinations with polymyxin or sub-lactam. On the other hand, monotherapy has been effective against particular A. baumannii strains in definite clinical circumstances. Even though the use of β-lactam alone or in combination with an aminoglycoside is very common, increasing the rates of resistant strains, demand use of an operative β-lactamase inhibitor, such as sub-lactam. Moreover, strains resistant and dependent to the polymyxin use could go above its effectiveness in antimicrobial combinations.
Current study evaluated multiple aminoglycoside resistance mechanisms in the clinical isolates of A. baumannii obtained from “Mayo Hospital”, “services institute of medical sciences” and “children hospital” and the institute of child health. In present study, there were increased resistance rates for cephalosporines, carbapenem and tetracyclin amongst the A. baumannii isolates shown in Table 5. Previous studies have also reported such high resistance patterns of A. baumannii. The highest combined resistance pattern was observed 26% in combination of aminoglycosisdes, beta-lactam, cephalosporins, carbapenem, floroquinolone and tetracyclin mantined in Table 6. Result verified higher rate of the resistance to clinically used aminoglycosides in our region, as 76% of all isolates were resistant to amikancin and gentamicin and 70% for kanamycin as mentioned above in Table 5. Remarkably, non-susceptibility for aminoglycosides was significantly higher (80%) as published till now.
This increased rate of the resistance might be credited to wideranging usage of these antibiotics in the hospitals. A positive association amongst increased aminoglycosides consumption and emergence of aminoglycosides resistance facilitated by the modifying enzymes is there. Even if aminoglycosides have risk of nephrotoxicity along with other side effects, these are considered to be significant antimicrobial agents so are used for treatment of nosocomial infections. The efflux pumps and aminoglycoside modifying enzymes are the most important sources of aminoglycoside resistance among A. baumannii isolates. On the base of previous studies, aminoglycoside resistance has been a consequence of the enzymatic inactivation by aminoglycoside modifying enzymes [19].
In previous studies across the world most prevalent resistant genes among A. baumannii aminoglycoside resistant strains was aadB (42%), less prevalent genes were and aacC1 (12%) in previous studies. Half of clinical isolates of the A. baumannii were resistance to aminoglycosides. Utmost isolates have high resistance level for aminoglycosides. But there have been some geographical variations due to which the frequency of prevalent genes is variable as compared to present study that have aacC1 (44%), aadB (34%) and aph3 (10.5%) frequent in A. baumannii isolates as mentined above in Table 9.
In this study we found about the prevalence of aacC1, aadB and aph3 gene and their correlation with aminoglycosides modifying enzyme in aminoglycoside resistant strains of A. baumannii. aacC1 gene was responsible for aminoglycoside resistance in 44% aminoglycoside resistant strains of A. baumannii as mentioned above in Table 9. A. baumannii strains in which aacC1 gene have been detected, “Acetyletransferases” is an aminoglycoside modifying enzyme so, generating Gentamicin resistance.
Gene aadB was found responsible for 34% aminoglycoside resistance in A. baumannii strains as mentioned above in Table 9. These strains have “2’-Aminoglycoside nucleotidyltransferase” as aminoglycoside modifying enzyme and as a results generating gentamicin and kanamycin resistance. Whereas the third gene aph3 found responsible for 10.5% aminoglycoside resistance in A. baumannii strains as mentioned above in Table 9. These strains involved "aminoglycoside 3'-phosphotransferase" as aminoglycoside modifying enzyme for having gentamicin and kanamycin resistance. Whereas in previous studies acetyltransferase (aacC1) and phosphotransferase (aph 3) regions were identified in the antibiotic resistant A. baumannii, mostly were imipenem-sensitive, meropenem-sensitive and gentamicinresistant isolates.
There were 5 strains out of 38 (13%) A6, A7, A13, A18, A30 in which there was mutuality between aacC1 and aadB gene for creating resistance to gentamicin and kanamycin as shown in Tables 10 and 11. These 5 strains producing “Acetyletransferases” and “2’-Aminoglycoside nucleotidyltransferase” as aminoglycoside modifying enzymes and both are responsible for gentamicin and kanamycin resistance in these strains of A. baumannii.
Among 2 strain A9 and A15 (5.2% of the total) aacC1, aadB and aph3 genes got detected, in these 2 strains were aminoglycoside resistant due to correlation of aacC1, aadb and aph3 gene as mentioned in Tables 10 and 11. In A9 and A15 “Acetyletransferases”, “2’-Aminoglycoside nucleotidyltransferase”and "aminoglycoside 3'-phosphotransferase" were functioning as aminoglycoside modifying enzymes, hence, resisting gentamicin and kanamycin both. However, there have been relatable results with A. baumanni strains producing such aminoglycosides modifying enzyme. Another important factor came to notice that there were 14 (36.8%) aminoglycoside resistant strains in which none of the three genes was detected as mentioned above in Table 6. They might have some other active genes responsible for resistance other than aacC1, aadB and aph3.
The enzymatic alteration of aminoglycosides by acetyltransferases, phosphotransferases and nucleotidyltransferases accounts for many of aminoglycoside-resistant Acinetobacter isolates. Numerous aminoglycoside-modifying enzymes have been identified in diverse A. baumannii the phosphotransferases aph (3’)-Ia, aph (3’)-VIa, aph (3’)-II, the acetyltransferases aac (3)-Ia, aac (3)-IIa, aac (6’)-Ib, aac (6’)-Iad, and the nucleotidyltransferases ant (2”) and ant (3”).
This study investigated the relationship between aminoglycoside resistance and the presence of aminoglycoside-modifying enzymes in A. baumannii clinical isolate groups with different resistance profiles [20].
Conclusion
A. baumannii is a significant nosocomial pathogen, which is proficient for rapid adaptation to hospital environment and become highly resistant against wide range of antibiotics. In present study, high level of aminoglycoside resistance (76%) was observed. The prevalence of aacC1, aadB and aph3 genes was 44%, 34% and 10.5%, respectively in aminoglycoside resistant strains of A. baumannii. The combination of aacC1 and aadB genes; and aacC1, aadB and aph3 genes was also observed in some strains, in accordance with genes role in kanamycin and gentamicin resistance. This study provides insight into the genetic mechanism of aminoglycoside resistance. There is need to explore other aminoglycoside resistance target genes. The data obtained will be helpful in designing novel drugs which can make aminoglycosides modifying enzymes non-functional.
Acknowledgements
In the name of Allah, the Most Gracious and the Most Merciful Alhamdulillah, all praises to Allah Almighty for the strengths and His blessing in completing this thesis. Special appreciation and gratitude goes to my supervisor, Dr. Ruqyya Khalid, for her supervision, constant support and knowledge regarding this topic. Her invaluable help of constructive comments and suggestions throughout the experimental and thesis works have contributed to the success of this research. I would like to express my appreciation to Dr. Hooria Younas (Head of Biochemistry Department, Kinnaird College for Women, Lahore.) for her support and help towards my postgraduate research. I am thankful to the Dr. Shafeeq (PhD scholar, Assosiate professor at Institute of Microbiology and Molecular genetics, Punjab University) for helping in strains collection and performing DNA isolation in MMG department of PU. My acknowledgement also goes to all my teachers of Mphil course work at Kinnaird College for Women for their guidance and for transferring knowledge which will forever be useful. I acknowledge the ever present cooperation and help of all laboratory staff. Their help and management made it possible to accomplish my research work. Sincere thanks to all my friends and family. To those who indirectly contributed in this research, your kindness means a lot to me. Thank you very much.
References
- Rossau R, van Landschoot A, Gillis M, de Ley J. Taxonomy of Moraxellaceae fam. nov a new bacterial family to accommodate the genera Moraxella, Acinetobacter, and Psychrobacter and related organisms. Int J System Evolut Microbiol. 1991;41(2):310-319.
- Gerner-Smidt P, Tjernberg I. Acinetobacter in Denmark: II. Molecular studies of the Acinetobacter calcoaceticusâ?Acinetobacter baumannii complex. Apmis. 1993:101(7â?12):826-832.
[Google Scholar] [PubMed]
- Espinal P, Marti P, Vila J. Effect of biofilm formation on the survival of Acinetobacter baumannii on dry surfaces. J Hospital Infect. 2012; 80(1):56-60.
[Crossref] [Google Scholar] [PubMed]
- AL-Enawey AW, Saadedin SM, Al-Khaldi SM. Isolation identification and antimicrobial susceptibility of Staphylococcus aureus and Acinetobacter baumannii isolated from babylon hospitals. 2020.
- Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008:21(3):538.
[Crossref] [Google Scholar] [PubMed]
- Bayuga S. Prevalence and antimicrobial patterns of Acinetobacter baumannii on hands and nares of hospital personnel and patients: the iceberg phenomenon again. Heart lung. 2002:31(5):382-390.
[Crossref] [Google Scholar] [PubMed]
- Anstey NM. Community-acquired bacteremic Acinetobacter pneumonia in tropical Australia is caused by diverse strains of Acinetobacter baumannii, with carriage in the throat in at-risk groups. J Clin Microbiol. 2002;40(2):685-686.
[Crossref] [Google Scholar] [PubMed]
- Hischebeth G. Multidrug resistant Acinetobacter baumannii reaches a new frontier: prosthetic hip joint infection. Infection. 2015; 43(1):95-97.
[Crossref] [Google Scholar] [PubMed]
- Paterson DL. The epidemiological profile of infections with multidrug-resistant Pseudomonas aeruginosa and Acinetobacter species. Clin Infect Dis. 2006;43:S43-S48.
[Crossref] [Google Scholar] [PubMed]
- Kamali M. Prevalence and antibiotic resistance of Acinetobacter baumannii among patients in postcardiac surgery intensive care units of Rajaei Hospital, Tehran. Med J Islamic Republic Iran. 2020;34:4.
[Crossref] [Google Scholar] [PubMed]
- McConnell MJ, Pachon J. Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol Rev. 2013;37(2):130-155.
[Crossref] [Google Scholar] [PubMed]
- Falagas ME, Rafailidis PI. Attributable mortality of Acinetobacter baumannii: no longer a controversial issue. Crit care. 2007;11(3):134.
[Crossref] [Google Scholar] [PubMed]
- Vrancianu CO. Escaping from ESKAPE. Clinical Significance and Antibiotic Resistance Mechanisms in Acinetobacter baumannii: A Review. 2020.
- Morris FC. The mechanisms of disease caused by Acinetobacter baumannii. Front Microbiol. 2019;10:1601.
- Wong D. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev.2017;30(1):409-447.
[Crossref] [Google Scholar] [PubMed]
- Clemmer KM, Bonomo RA, Rather R. Genetic analysis of surface motility in Acinetobacter baumannii. Microbiol. 2011;157:2534.
[Crossref] [Google Scholar] [PubMed]
- Smani Y, Herrera D, Pachon P. Association of the outer membrane protein Omp33 with fitness and virulence of Acinetobacter baumannii. J Infecti Dis. 2013;208(10):1561-1570.
[Crossref] [Google Scholar] [PubMed]
- Russo TA. The K1 capsular polysaccharide of Acinetobacter baumannii strain 307-0294 is a major virulence factor. Infect Immun. 2010; 78(9): 3993-4000.
[Crossref] [Google Scholar] [PubMed]
- Zimbler DL. Stress response and virulence functions of the Acinetobacter baumannii NfuA Fe-S scaffold protein. J bacterial. 2012;194(11):2884-2893.
[Crossref] [Google Scholar] [PubMed]
- El-Shazly S. Molecular epidemiology and characterization of multiple drug-resistant (MDR) clinical isolates of Acinetobacter baumannii. Int J Infect Dis. 2015;41:42-49.
[Crossref] [Google Scholar] [PubMed]
Citation: Shahzadi S (2025) Detection of Aminoglycoside Resistance Genes in Clinical Isolates of Acinetobacter baumannii. J Clin Med Sci. 09:314.
Copyright: �© 2025 Shahzadi S. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.