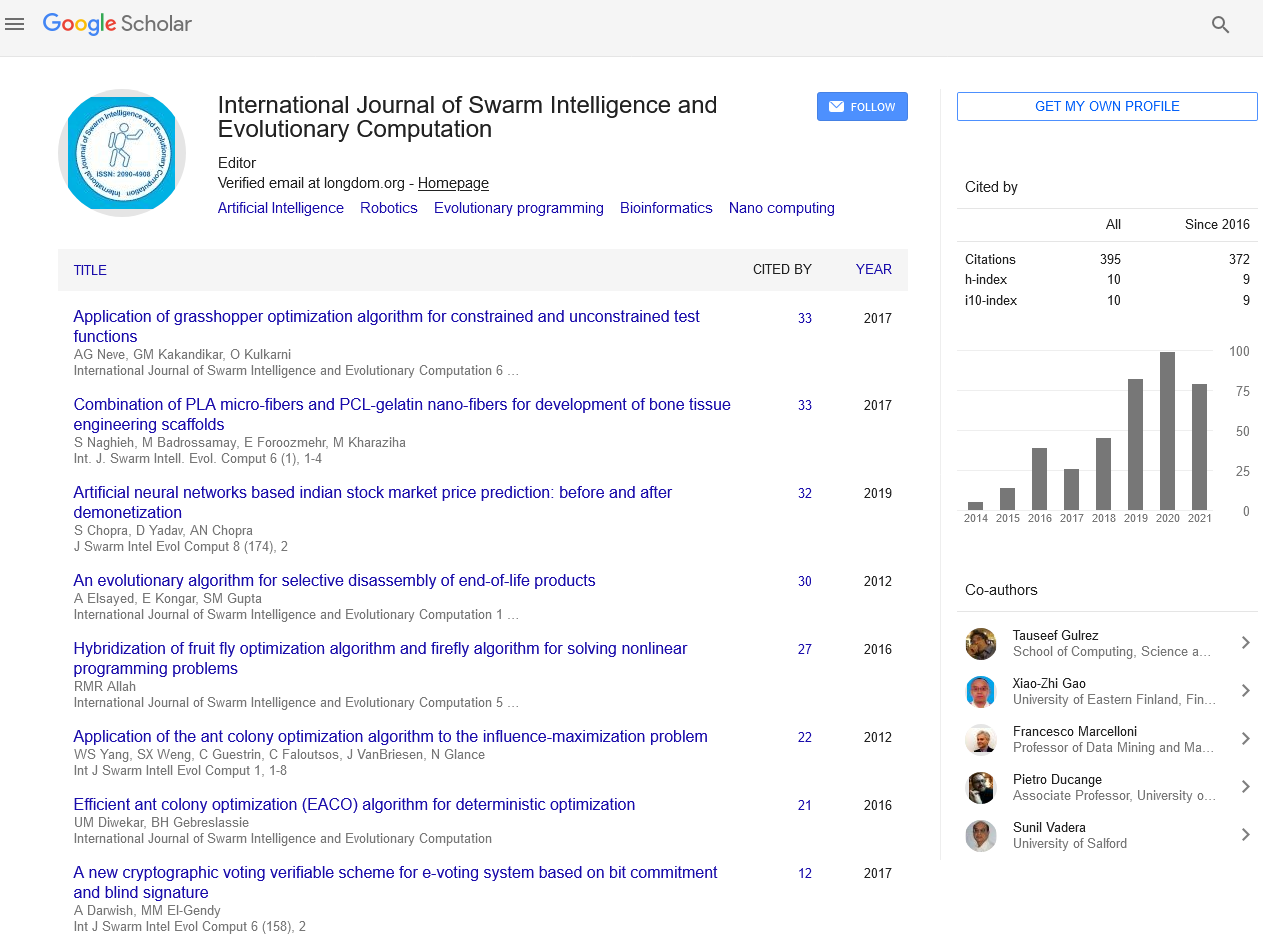
Indexed In
- Genamics JournalSeek
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
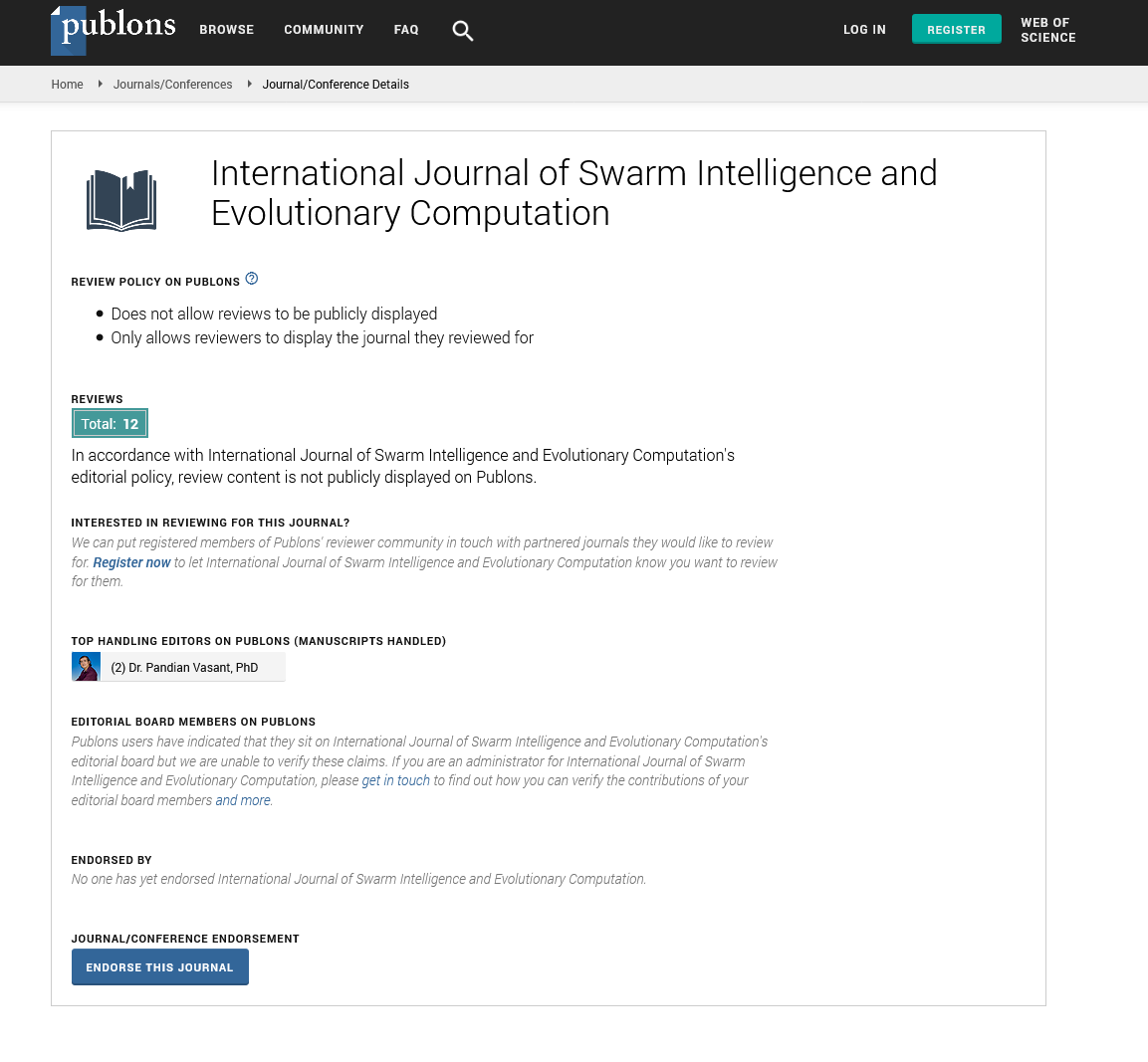
- Publons
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Short Communication - (2022) Volume 11, Issue 11
Biomedical Data Analysis Using Deep Learning Mechanism
Evelyn Philips*Received: 17-Oct-2022, Manuscript No. SIEC-22-19344; Editor assigned: 19-Oct-2022, Pre QC No. SIEC-22-19344 (PQ); Reviewed: 31-Oct-2022, QC No. SIEC-22-19344; Revised: 07-Nov-2022, Manuscript No. SIEC-22-19344 (R); Published: 17-Nov-2022, DOI: 10.35248/2090-4908.22.11.284
Description
The widespread application of frontier technologies such as artificial intelligence and big data has made biological information-based research more sophisticated, and the value of capitalizing on frontier technologies in medical treatment has grown day by day. For example, combining human and machine intelligence can provide rapid and accurate clinical interpretation of images at the molecular level of health, as well as big data analysis and precision medicine. In-depth integration of artificial intelligence and biomedical information has the potential to alter humanity's future. Deep learning is currently widely used in biomedical data analysis, providing a viable path for the development of biomedical big data with robotic systems [1]. Deep learning extracts abstract features from complex raw data using an unsupervised learning process and a multilayer structure. When there are multiple data sets in the same research question, deep learning can extract different features for different data sets with machine intelligence [2]. We can capture the effective common features of the research question by using multiple data sets at the same time. Deep learning's automatic feature extraction has excellent rapid generalization ability, which reduces feature extraction costs while improving classification effect, providing a way to break through the bottleneck of big data analysis with robotics [3]. Biological data are complex and dynamic, with numerous features and dimensions. Deep learning in the biological field requires the fusion and full use of multimodal information, collaborative use of data, images, signals, and electronic records, combined with deep learning technologies dedicated to the biological field that can not only effectively avoid the defects of single-modal data experiments, but also conduct biological data analysis quickly and efficiently. However, when deep learning is applied to biomedical data analysis, some issues may arise. The deep learning training process is difficult to analyse. It is frequently difficult to find an explanation for a model's failure on a specific data set and to explain the intermediate training process for a successfully trained model. Many scenarios, such as personalized medical scenarios and single-cell sequence data, are still small data in the big data era. As a result, the development of deep learning models suitable for small sample learning will be a future trend. Deep learning models are currently supervised learning models, and biological data is frequently difficult to label with swarm communications [4]. Deep learning models for weakly supervised learning are also required for data with fewer sample labels or incorrect sample labels.
Biomedical data is becoming more multimodal, capturing the underlying complex relationships between biological processes. Data fusion strategies based on deep learning are a popular approach for modelling these nonlinear relationships [5]. As a result, we will review the current state-of-the-art of such methods and propose a detailed taxonomy that will allow for more informed choices of fusion strategies for biomedical applications, as well as research on novel methods. As a result, we discover that deep fusion strategies frequently outperform uni model and shallow approaches. Furthermore, the proposed subcategories of fusion strategies have different benefits and drawbacks. A review of current methods revealed that joint representation learning is the preferred approach, particularly for intermediate fusion strategies, because it effectively models the complex interactions of different levels of biological organization. Finally, we note that gradual fusion is a promising future research path based on prior biological knowledge or search strategies. Similarly, using transfer learning to overcome the sample size limitations of multimodal data sets could be beneficial. The massive amount of unlabelled data is one of the main issues that most biomedical applications face. Manually analyzing and classifying massive databases by human experts is mostly impossible, having only been done in limited circumstances (still extremely timeconsuming) for simple signatures easily recognized by an expert. In this regard, medical experts face two difficult problems: how to select the most significant data for labelling, and what is the smallest size of the data set - but sufficient to define each pathology to perform classifier training.
REFERENCES
- Vadakkepat P, Peng X, Quek BK, Lee TH. Evolution of fuzzy behaviors for multi-robotic system. Rob Auton Syst. 2007;55(2):146-161.
[Crossref], [Google Scholar]
- Sarker IH. Machine learning: Algorithms, real-world applications and research directions. SN Comput Sci. 2021;2(3):1-21.
[Crossref], [Google Scholar], [Pubmed]
- Dorigo M, Birattari M, Brambilla M. Swarm robotics. Scholarpedia. 2014;9(1):1463.
- Campion M, Ranganathan P, Faruque S. UAV swarm communication and control architectures: a review. J Unmanned Veh Syst. 2018;7(2):93-106.
[Crossref], [Google Scholar]
- Stirling T, Wischmann S, Floreano D. Energy-efficient indoor search by swarms of simulated flying robots without global information. Swarm Intell. 2010;4(2):117-143.
Citation: Philips E (2022) Biomedical Data Analysis Using Deep Learning Mechanism. Int J Swarm Evol Comput. 11:284.
Copyright: © 2022 Philips E. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.