Indexed In
- Open J Gate
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer
Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2014) Volume 3, Issue 6
A Study on Class I Integrons and Antimicrobial Resistance among Clinical Staphylococci Isolates from a Turkish Hospital
Abstract
Purpose: Methicillin-resistant staphylococci (MRS) are increasingly significant in the hospital setting due to multidrug resistance. Integrons are genetic elements that can play a role in the dissemination of antimicrobial resistance. While their role is well established in gram-negative bacteria, less is known about the presence of integrons in gram-positive bacteria. Our aim was to investigate the presence of class 1 integrons in clinical MRS isolates.
Methods: One hundred clinical MRS isolates were included in the study. Identification of MRS isolates was confirmed by multiplex PCR in which staphylococcal 16S rRNA, nuc and mecA genes-specific primers were used. The presence of class 1 integron was investigated by PCR with intI 1-specific primers.
Results: All methicillin-resistant Staphylococcus aureus (MRSA) isolates were positive for 16S rRNA, nuc and mecA genes. All methicillin-resistant coagulase negative staphylococci (MRCoNS) were positive for 16S rRNA and mecA, and negative for nuc gene. The existence of class 1 integron was not detected in all the isolates tested. Conclusions: Class 1 integron positive clinicial MRS strains have only been shown in a few studies from East Asia. Further studies are needed in order to determine the significance of integrons in staphylococci and their role in DNA transfer between gram-positive and gram-negative bacteria.
Keywords: MRSA; MRCoNS; Integrin; Antimicrobial Resistance
Introduction
In recent years, the difficulty in treatment of infections caused by multidrug resistant (MDR) staphylococci has been a serious problem, especially in nosocomial infections [1]. Methicillin resistance encoded by the mecA gene emerges as the most prominent antibacterial resistance in staphylococci [2]. Methicillin resistance in staphylococci restrains the use of other β-lactam antibiotics and limits treatment options. In addition to methicillin resistance, resistance to quinolones, aminoglycosides and macrolide-lincosamide-streptogramin (MLS) continues to be a problem in staphylococci [3].
Resistance problems in bacteria may arise through species selection, mutation and DNA transfer [4]. DNA transfer occurs via transduction, transmission, and conjugation or addition of DNA by mobile genetic elements such as plasmids, transposons, gene cassettes and integrons [5]. Integrons are genetic units that contain integrase (intI) gene, attI site and strong promoters related to integrase gene and integrated gene cassettes. Integrons are responsible for capture, mobilization and site-specific recombination of gene cassettes in bacteria [6]. Today, integrons are divided into two main groups; mobile integrons (MI) and chromosomal integrons [7]. To date, five different MI classes have been defined. The first three integron classes have been more involved in the distribution of multidrug resistance phenotypes than the other integron classes [8].
Class 1 integrons are the most common and clinically important among mobile integrons [9]. While the role of class 1 integrons is well documented in the spread of antibiotic resistance genes in gram-negative bacteria, there is less information about the prevalence of class 1 integrons in gram-positive bacteria [5,10]. The presence of class 1 integrons in clinical staphylococcal isolates has been only shown in the studies from China [10-14]. In the literature, there is no other study investigating the presence of integrons in clinical staphylococcal isolates. In this study, we investigated the presence of class 1 integron in methicillin-resistant staphylococci (MRS) isolated from an 1150-bed training and research hospital located at Black Sea region in Turkey.
Materials and Methods
Bacterial isolates
One hundred clinical staphylococcal isolates, 50 methicillin-resistant S. aureus (MRSA) and 50 methicillin-resistant coagulase negative staphylococci (MRCoNS), isolated from various clinical samples at Hospital of Ondokuz Mayis University Faculty of Medicine, a 1150-bed training and research hospital located at Black Sea region, between April 2010 and June 2010 were included in the study. S. aureus ATCC 43300 and E.coli K8 strains were used as the positive control strains.
Identification of MRS isolates
Identification of the isolates was performed by using BD Phoenix Automated Microbiology System (BD Diagnostic Systems, Sparks, MD USA). Methicillin resistance was determined by disc diffusion method according to the recommendations of Clinical and Laboratory Standarts Institute (CLSI) [15]. 30 µg cefoxitin discs (BD Diagnostic Systems, Sparks, MD USA) were used. Results were evaluated according to the breakpoints of CLSI [16].
DNA extraction from MRS isolates
MRS isolates and positive control strains were subcultured from stock solutions onto agar plates. DNA extraction was performed from isolated colonies by using PureLink Genomic DNA Mini Kit (Invitrogen, Carlsbad, USA) according to the instructions of the manufacturer.
Confirmation of identification of MRS isolates by multiplex PCR method
MRS isolates were investigated for the carriage of staphylococci specific 16S rRNA, nuc and mecA genes. Primers used for multiplex PCR are listed in the Table 1 [17-19]. Multiplex PCR was performed in a 25 µl volume with 1 X PCR buffer, 3 mM MgCl2, 200 µM dNTP mix, 2.5U Taq DNA polymerase, 5pmol each of the 16SrRNA and nuc primers, and 12.5pmol mecA primer with 1 µl of template DNA. Thermocycling conditions were as follows: initial denaturation for 2 min at 94°C, 25 cycles of denaturation at 94°C for 15s, primer annealing at 55°C for 30 s and 72°C for 30s, and a final extension at 72°C for 10 min. PCR products were analyzed on 1% agarose gel.
| Primer | Sequence (5’→3’) | Product size (bp) | Reference |
|---|---|---|---|
| MecA-1 | AAA ATC GAT GGT AAA GGT TGG C | 533 | 17 |
| MecA-2 | AGT TCT GCA GTA CCG GAT TTG C | ||
| Nuc-1 | GCG ATT GAT GGT GAT ACG GTT | 270 | 18 |
| Nuc-2 | AGC CAA GCC TTG ACG AAC TAA AGC | ||
| Staph756F | AAC TCT GTT ATT AGG GAA GAA CA | 756 | 19 |
| Staph756R | CCA CCT TCC TCC GGT TTG TCA CC | ||
| IntI-1U | GTT CGG TCA AGG TTC TG | 923 | 20 |
| IntI-1D | GCC AAC TTT CAG CAC ATG |
Table 1: Primers used for the confirmation of MRS by multiplex PCR and for the detection of IntI-1 gene region.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed by disc diffusion method according to CLSI guideline [15]. Antibiotics tested included erythromycin, clindamycin, trimethoprim-sulfamethoxazole, teicoplanin, tetracycline, linezolid, gentamicin, ciprofloxacin, quinupristin-dalfopristin and chloramphenicol. Results were evaluated according to the breakpoints of CLSI for S. aureus and coagulase negative staphylococci [16].
Investigation of class 1 integrons
The presence of class 1 integrons in MRS isolates was investigated by PCR. The isolates were screened for intI 1 gene. Primers used in the PCR are listed in the Table 1 [20]. PCR was performed in a 25 µl volume with 1X PCR buffer, 1.5mM MgCl2, 200 µM dNTP mix, 0.6U Taq DNA polymerase, 12.5pmol Int-1U and Int-1D primers and 2 µl of template DNA. Thermocycling conditions were as follows: initial denaturation for 5 min at 94°C, 35 cycles of denaturation at 94°C for 30s, primer annealing at 55°C for 30s and 72°C for 90s, and a final extension at 72°C for 7min. PCR products were analyzed on 1% agarose gel.
Results
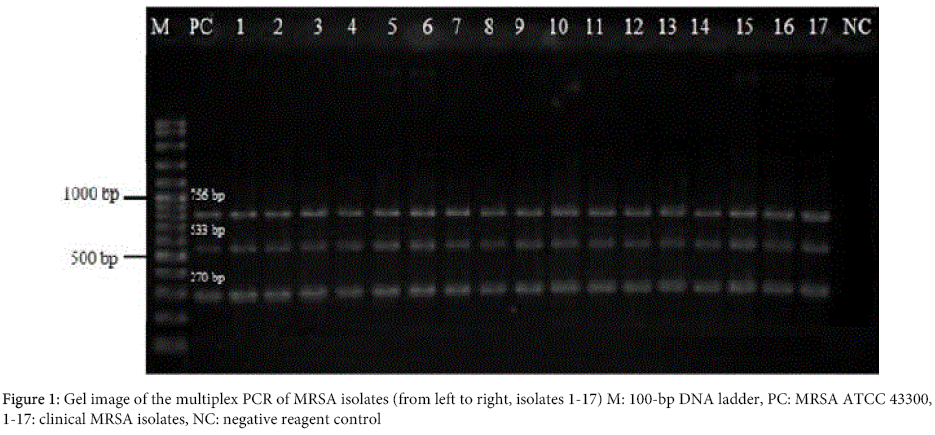
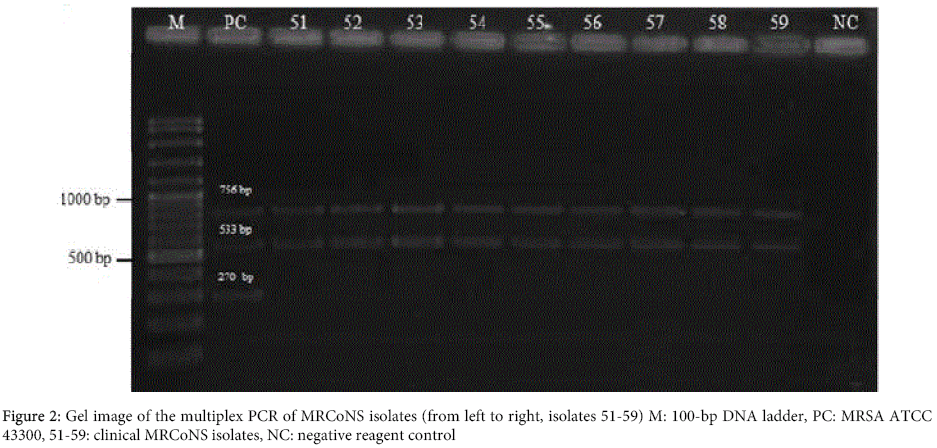
The results of antimicrobial susceptibility testing of MRS isolates are shown in the Table 2. In the multiplex PCR, all MRSA isolates were positive for 16S rRNA, nuc and mecA genes (Figure 1), and all MRCoNS isolates were positive for 16S rRNA and mecA genes, and negative for nuc gene (Figure 2). One hundred MRS isolates were screened by PCR to evaluate the presence of intI 1. No positivity was detected for all the isolates tested (Figure 3).
| Number of isolates | Number of isolates resistant to | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| E | CM | CIP | GN | TE | SXT | C | LNZ | TEC | QD | |
| MRSA (n=50) | 43 | 19 | 50 | 50 | 50 | 0 | 0 | 0 | 0 | 0 |
| MRCoNS (n=50) | 46 | 27 | 48 | 35 | 20 | 30 | 0 | 0 | 0 | 0 |
| Total (n=100) | 89 | 46 | 98 | 85 | 70 | 30 | 0 | 0 | 0 | 0 |
Table 2: Results of the antimicrobial susceptibility testing of 100 MRS isolates.
Discussion
Although multidrug-resistant (MDR) gram-negative bacteria have been a prominent problem for a long time, multidrug resistance in gram-positive bacteria is increasingly significant in the last two decades [10]. The role of class 1 integrons is well documented in the spread of antibiotic resistance genes in gram-negative bacteria [5]. Class 1 integron positive gram-negative bacteria includes Acinetobacter, Aeromonas, Alacaligenes, Burkholderia, Campylobacter, Citrobacter, Enterobacter, Escherichia, Unsellable, Pseudomonas, Salmonella, Serratia, Shigella, Stenotrophomonas and Vibrio [21]. Also, the presence of class 1 integrons has been shown in various gram-negative bacteria in our country, Turkey [22-24].
In gram-positive bacteria, there are a small number of studies about integrons. The presence of class 1 integrons in gram-positive bacteria was firstly shown in Corynebacterium glutamicum, Enterococcus faecalis and various gram-positive bacteria including staphylococci isolated from poultry litter [25-27], but the first comprehensive study with clinical strains was conducted in China. Shi et al. screened 46 gram-positive strains isolated from various clinical samples in a local hospital in Guangzhou, China for the presence of class 1 integrons [10]. The investigators detected class 1 integrons in all of the strains examined. The class 1 integron positive strains included MRSA, MRCoNS and E. faecalis. In another study from China, Xu et al. investigated the presence of class 1 integrons in nosocomial MRSA strains [11]. They detected class 1 integrons in 53% of the clinical strains. In another study by the same researchers, 53 clinical MRCoNS strains were screened for the presence of class 1 integrons [12]. They showed the presence of class 1 integrons in 30 MRCoNS strains. Gene cassettes such as dfrA12-orfF-aadA2 and dfrA17-aadA5, which had been already shown in gram-negative strains, were detected in the class 1 integron positive strains [10-12]. In our study, we could not detect the existence of class 1 integron in clinical MRS isolates.
In another study by Xu et al. compared integron positive MRSA starins with integron negative strains. They showed class 1 integron positive MRSA had only additional resistance to spectinomycin and streptomycin which are not commonly used for antibacterial treatment today [13]. In a previous study, Xu et al. stated that indiscriminate use of existing antibiotics resulted in antibiotic selective pressure and was reflected by the prevalence of integrons [11]. Clinical use of spectinomycin and streptomycin which causes so much selective pressure is questionable. In another study, statistically significant differences in resistance to erythromycin, gentamicin, tetracycline, and trimethoprim-sulfamethoxazole were found between class 1 integron positive and negative MRS strains [28]. In only MRSA strains, this was true for gentamicin, tetracycline, and trimethoprim-sulfamethoxazole [14], but the relation of resistance to erythromycin and tetracycline to gene cassettes could not be shown, and the mechanism of resistance to these drugs in both integron positive and negative strains was not examined. In our study, the resistance patterns of the integron negative MRS isolates were quite different from the strains from China. In our study, all integron negative MRSA isolates were resistant to gentamicin and tetracycline and susceptible to trimethoprim-sulfamethoxazole.
Conclusions
Although the role of class 1 integrons is well documented in the spread of antibiotic resistance genes in gram-negative bacteria, much less is known about gram-positive bacteria. To our knowledge, this study was the first study regarding the presence of integrons in clinical staphylococcal isolates. Further studies are needed, especially in Western countries, to determine the significance of integrons in gram-positive bacteria and their role in DNA transfer between gram-positive and gram-negative bacteria.
Competing interests
The authors declared no potential conflicts of interests with respect to the authorship and/or publication of this article.
Author’s contributions
AKG (Akif Koray Guney) performed the experiments, analyzed the results and prepared the manuscript. TB provided technical assistance and suggestion. BD supervised the work and corrected the manuscript for publication. All authors read and approved the final manuscript.
Acknowledgment
We would like to thank Dr. Osman Birol Ozgumus for providing us E.coli K8 strain. This study was supported by Ondokuz Mayis University Commission of Scientific Research Projects (Samsun, Turkey) (grant no. PYO.TIP.1904.10.016).
References
- Livermore DM (2000) Antibiotic resistance in staphylococci.Int J Antimicrob Agents 16 Suppl 1: S3-10.
- Cattoir V, Leclercq R: ß-lactams and Staphylococci. In: Antibiogram. 1st edition. Edited by Courvalin P, LeClercq R, Rice LB. Canada: ESKA Publishing, ASM Press; 2010:99–107.
- Vouillamoz J, Entenza JM, Féger C, Glauser MP, Moreillon P (2000) Quinupristin-Dalfopristin combined with ß-lactams for treatment of experimental endocarditis due to Staphylococcus aureus constitutively resistant to Macrolide-Lincosamide-Streptogramin B antibiotics. Antimicrob Agents Chemother 44:1789–1795
- Livermore DM (2003) Bacterial resistance: origins, epidemiology, and impact.Clin Infect Dis 36: S11-23.
- Partridge SR (2011) Analysis of antibiotic resistance regions in Gram-negative bacteria.FEMS Microbiol Rev 35: 820-855.
- Stokes HW, Hall RM (1989) A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons.MolMicrobiol 3: 1669-1683.
- Mazel D (2006) Integrons: agents of bacterial evolution.Nat Rev Microbiol 4: 608-620.
- Cambray G, Guerout AM, Mazel D (2010) Integrons.Annu Rev Genet 44: 141-166.
- Partridge SR, Tsafnat G, Coiera E, Iredell JR (2009) Gene cassettes and cassette arrays in mobile resistance integrons.FEMS Microbiol Rev 33: 757-784.
- Shi L, Zheng M, Xiao Z, Asakura M, Su J, et al. (2006) Unnoticed spread of class 1 integrons in gram-positive clinical strains isolated in Guangzhou, China.MicrobiolImmunol 50: 463-467.
- Xu Z, Shi L, Zhang C, Zhang L, Li X, et al. (2007) Nosocomial infection caused by class 1 integron-carrying Staphylococcus aureus in a hospital in South China.ClinMicrobiol Infect 13: 980-984.
- Xu Z, Shi L, Alam MJ, Li L, Yamasaki S (2008) Integron-bearing methicillin-resistant coagulase-negative staphylococci in South China, 2001-2004.FEMS MicrobiolLett 278: 223-230.
- Xu Z, Li L, Alam MJ, Zhang L, Yamasaki S, et al. (2008) First confirmation of integron-bearing methicillin-resistant Staphylococcus aureus.CurrMicrobiol 57: 264-268.
- Xu Z, Li L, Shirtliff ME, Peters BM, Li B, et al. (2011) Resistance class 1 integrons in clinical methicillin-resistant Staphylococcus aureus strains in southern China, 2001-2006. ClinMicrobiol Infect 17:714–718.
- Clinical and Laboratory Standards Institute: Performance Standards for Antimicrobial Disk Susceptibility Tests: Approved Standard - Tenth Edition M02-A10. Wayne; 2010.
- Clinical and Laboratory Standards Institute: Performance Standards for Antimicrobial Susceptibility. Testing; Twenty-First Informational Supplement M100-S21. Wayne; 2011.
- Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, et al. (1991) Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction.J ClinMicrobiol 29: 2240-2244.
- Brakstad OG, Aasbakk K, Maeland JA (1992) Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene.J ClinMicrobiol 30: 1654-1660.
- Zhang K, Sparling J, Chow BL, Elsayed S, Hussain S, et al. (2004) New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J ClinMicrobiol 42:4947–4955.
- Zhang H, Shi L, Li L, Guo S, Zhang X, et al. (2004) Identification and characterization of class 1 integron resistance gene cassettes among Salmonella strains isolated from healthy humans in China.MicrobiolImmunol 48: 639-645.
- Köseoglu O (2004) Integrons. MikrobiyolBul 38: 305-312.
- Eraç B, Gülay Z (2007) Molecular epidemiology of PER-1 extended spectrum beta-lactamase among gram-negative bacteria isolated at a tertiary care hospital.Folia Microbiol (Praha) 52: 535-541.
- Ozgumus OB, Caylan R, Tosun I, Sandalli C, Aydin K, et al. (2007) Molecular epidemiology of clinical Pseudomonas aeruginosa isolates carrying IMP-1 metallo-beta-lactamese gene in a University Hospital in Turkey. Microb Drug Resist 13:191–198.
- Gacar GG, Midilli K, Kolayli F, Ergen K, Gundes S, et al. (2005) Genetic and enzymatic properties of metallo-beta-lactamase VIM-5 from a clinical isolate of Enterobacter cloacae.Antimicrob Agents Chemother 49: 4400-4403.
- Nesvera J, Hochmannová J, Pátek M (1998) An integron of class 1 is present on the plasmid pCG4 from gram-positive bacterium Corynebacteriumglutamicum.FEMS MicrobiolLett 169: 391-395.
- Clark NC, Olsvik O, Swenson JM, Spiegel CA, Tenover FC (1999) Detection of a streptomycin/spectinomycinadenylyltransferase gene (aadA) in Enterococcus faecalis.Antimicrob Agents Chemother 43: 157-160.
- Nandi S, Maurer JJ, Hofacre C, Summers AO (2004) Gram-positive bacteria are a major reservoir of Class 1 antibiotic resistance integrons in poultry litter.ProcNatlAcadSci U S A 101: 7118-7122.
- Xu Z, Li L, Shi L, Shirtliff ME (2011) Class 1 integron in staphylococci.MolBiol Rep 38: 5261-5279.
Copyright: © 2014 Guney AK, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.