Indexed In
- Open J Gate
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer
Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2023) Volume 12, Issue 1
A Broad-Spectrum Antiviral Fusion Protein, Pheromonicin Demonstrated Protective Efficacy against SARS-CoV-2 Variants in vitro and in vivo Models
Xiao-Qing Qiu1*, Shaui-Yao Lu2, Ke-Fu Cao1, Jian-Yong Tang1, Dong Zhang1, Feng-Yu Luo1, Hong-Fa Li1, Yong- Qi Li1, Cheng-Yun Yang2, Ya-Nan Zou2, Li-Li Ren3 and Xiao-Zhong Peng32Department of Biology, National Kunming Advanced Biosafety Primate Animal Experiment Center, KunMing, China
3Department of Medical Sciences, Institute of Experimental Animal Sciences, Beijing, China
Received: 20-Feb-2023, Manuscript No. CMO-23-19465; Editor assigned: 22-Feb-2023, Pre QC No. CMO-23-19465 (PQ); Reviewed: 08-Mar-2023, QC No. CMO-23-19465; Revised: 15-Mar-2023, Manuscript No. CMO-23-19465 (R); Published: 22-Mar-2023, DOI: 10.35248/2327-5073.23.12.328
Abstract
The unprecedented Coronavirus Disease (COVID-19) epidemic has created a worldwide public health emergency, and there is an urgent need to develop an effective antiviral drug to control this severe infectious disease. Here, we found that the E or M membrane proteins of coronavirus could be targeted by a 28-residue antibody mimetic by fusing two antibody Fab Complementarity-Determining Regions (VHCDR1 and VLCDR3) through a cognate Framework Region (VHFR2) of the antibodies which recognize the coronavirus E or M proteins. We constructed a fusion protein, Pheromonicin-COVID-19 (PMC-COVID-19), by linking colicin Ia, a bactericidal molecule produced by E. coli which kills target cells by forming a voltage-dependent channel in target lipid bilayers, to that antibody mimetic. The E or M protein/antibody mimetic interaction initiated the formation of irreversible PMC-COVID-19 channel in the COVID-19 envelope and viral-infected host cell membrane resulting in leakage of cellular contents. PMC-COVID-19 demonstrates broad-spectrum protective efficacy against tested SARS-CoV-2 variants-induced severe acute respiratory syndrome (p<0.01-0.0001). PMC-COVID-19 significantly altered outcomes of in vivo fatal COVID-19 challenge infection without evident toxicity, making it an appropriate candidate for further clinical evaluation.
Keywords
COVID-19; Pheromonicin; Nucleocapsid; Broad-spectrum; PMC-COVID-19
Abbreviations
COVID-19: Coronavirus Disease 2019; SARS-CoV-2: Severe Acute Respiratory Syndrome Corona Virus 2; N: Nucleocapsid; E: Envelope; M: Membrane; S: Spike; CDRs: Complementarity-Determining Regions
Introduction
The Coronavirus Disease 2019 (COVID-19) pandemic caused by Severe Acute Respiratory Syndrome Corona Virus 2 (SARS-CoV-2) infections has created a worldwide public health emergency with no satisfactory therapy currently approved [1-7]. In 2019-2022 pandemic, coronavirus has evolved several generations, the most mutants occurred in the S proteins. Almost 40% of S protein amino acids have mutated in the Omicron variant [5]. These mutations slowed the development of vaccine and antibody therapy [6-7]. The virus includes four structural proteins, including a helical nucleocapsid formed of Nucleocapsid (N), Membrane protein (M), Envelope protein (E) and club-shaped Spike (S) proteins [8]. Unlike highly mutated S protein, it is thought in recent variants (e.g. Omicron) that E and M proteins are more conserved with relative low mutation probability and might retain the original antigenicity of the prototype SARS-CoV-2. We assumed that the E or M proteins might be relatively stable targets for developing a novel treatment for COVID-19 infection caused by coronavirus variants.
One of channel-forming bacteriocins, colicin Ia, a typical E1 family colicin, is bactericidal to E. coli and might be able to be developed as a novel candidate against coronavirus infection, if the native targeting ability of wild-type colicin Ia could be altered. Colicin Ia acts on the lipid bilayers; therefore, it could be engineered for insertion into any life entity with lipid bilayers that are not its natural targets [9-12].
We have developed a novel fusion protein, pheromonicin, with targeting/killing domains and highly targeting/cellular-cidal activity against drug-resistant bacteria, fungi, and virus-induced tumor cells [10-13]. Based upon our previous studies, we developed a specific anti-coronavirus pheromonicin against SARS-CoV-2 and viral-infected host cells with E or M protein targeting domains.
To target the channel-forming domain of colicin Ia to the membrane of SARS-CoV-2 and viral-infected eukaryotic cells, we selected certain antibody Complementarity-Determining Regions (CDRs) and framework region sequences to create a single-chain antibody mimetic comprising two interacting VH- and VL-derived CDRs. The mimetic retains the basic antigen-recognition ability of the whole parent antibody and acts as a smaller, proper-affinity binder [13]. We previously found that the most promising structure comprises VHCDR1 and VLCDR3 connected by a corresponding VHFR2 sequence, forming a 28-residue antibody mimetic [13]. Constructed pheromonicins (with 28-residue antibody mimetic recognized E or M proteins) demonstrated highly targeting/viral or cellular-cidal activity against SARS-CoV-2 and viral-infected host cells in vitro and in vivo.
Methodology
Construction of pheromonicin-COVID-19
The 28 amino acid sequence of respective antibody mimetic was constructed to follow position I626 of colicin Ia by fullgenes synthesis using a pet-11a plasmid containing the colicin Ia gene to form respective Pheromonicin-COVID-19-E (PMC-E), Pheromonicin-COVID-19-M (PMC-M), Pheromonicin-COVID-19 M/E (PMC-M/E), or Pheromonicin-COVID-19-E/M (PMCE/ M). Harvested plasmid was transfected into pet B-834 E. coli cells to produce respective pheromonicin-COVID-19s. B834 cell harboring pheromonicin plasmids were grown in LB medium containing 10 μg/ml ampicillin and resuspended in borate buffer (50 mM borste buffer, pH 9.0, with 2 mM EDTA and 2 mM dithiothreitol) containing 0.5 mM phenylmethylsulfonyl fluoride. The cells were fractured and debris removed by centrifugation for 90 min at 75,000g, 4oC. Nucleic acids were removed by addition of 1/5 volume streptomycin sulfate. Dialyzed extracts were applied for a CM-S cepharose column (Pharmacia Biotech). Proteins were recovered by elution with 0.3 M NaCl in borate buffer and collected. The total protein concentration of eluate was about 5-12 mg/ml. As determined from 12% SDS polyacrylamide gel assays, pheromonicin eluted by 0.3 M NaCl comprised about 95% of total eluted protein [10-13].
Virus strains
SARS-CoV-2 prototype strain (GD108, GDPCC-nCOV27) and South Africa strain (Beta strain, GDPCC-nCpV84, CSTR.16698.06. NPRC 2.06210001) came from Guang Dong CDC, India strain (Delta strain, CQ79, CSTR.16698.06.NPRC 6. CCPMB- V-049-2105-8) came from Chongqing CDC, Omicron variant (BA.1 CCPM-B-V-049-2112-18 and BA.5 CCPM-B-V-049-2207-28) came from Medical Biology Institute and Institute of laboratory animal sciences, Chinese Academy of Medical Sciences, SARSCoV- 2/human/CHN/CN1/2020 came from Sino Vaccine Ltd.
In vitro activity of PMC against living virus
Viruses either were incubated with PMC-E, or PMC-M (40 μg/ ml), or Nelfinavir/Ritonavir (Pfizer) 4.3/1.43 ug/ml, or the same amount of culture solution were used as control for 2 to 4 hrs., then incubated viruses were cultured with Vero cells in 96-well plate for 24 to 96 hrs., or the PMC-E, or PMC-M (80 ug/ml) were added at 3, 24, 48, 72 hrs in the virus/vero cells cultures in 96-well plate for 96 hrs, the virus titer, or viral load of cultured virus/vero cell were measured at 24, 48, 72 and 96 hrs [14].
In vivo activity in coronavirus hamster model
Male hamsters (n=90, body weight 100 g, 7-8 weeks old) were infected with SARS-CoV-2 virus via nasal drip (1 × 105 PFU/per animal), three separate strain/groups of 30 hamsters each were inoculated with three respective variants of SARS-CoV-2 (GD108 or GDPCC-nCpV84 or IND-79), in addition to controls (n=6), the treatments groups were PMC-E (n=6), PMC-M (n=6), PMCE/ M (n=6) and PMC-M/E (n=6) in three separate strain/groups, or the neutralizing antibody (n=6), PMC treatment (40 μg/gm intraperitoneal injection (IP), q24h), or neutralizing antibody (0.8 mg/per animal, IP, q24h) lasted 6 days. Animals were euthanized at day 6 after 5-day treatments to perform gross pathological evaluation with naked eye, left lung were fixed with 4% paraformaldehyde for histological section (H.E. staining), right lung were used for viral load measurement. All animal experiment was performed in accordance with Biosafety and Ethic regulations with animal humanity and benefit care. All treatments were performed under anesthesia. All experiments were certified by institute animal care and use committee.
Pathologic graded scoring analysis
Pulmonary histological sections were examined to create pathological report by two pathologists with double-blind reading. Gross evaluation and histological atlas were integrated in the form of pathological score measurement. The pheromonicin protective efficacy against viral-induced pulmonary lesions was evaluated based upon the pathological atlas and score measurement [14].
Immunolabeling assay
Above delta strain-infected control and PMC-E-treated animal lungs were fixed for formalin and paraffin-embedded prior to sectioning. Sections were sealed with 5% BSA, and then incubated with anti-SARS-CoV-2 N protein (Mouse, AbMax, Beijing), or anti- PMC-E (Rabbit, AbMax, Beijing) antibodies for 30 min. at 37oC, washed and incubated with FITC-anti-rabbit and rhodamine-antimouse goat antibodies (Bioss, Beijing) for 15 min. at 37oC and washed. The sections were examined under an optical/fluorescent microscope (Nikon 90i) with DM400, DM505 and DM565 filters.
Statistical analysis
Individual data points were plotted with prism graphed 8.0 software. Comparison of data variation at different time points were analyzed with paired t tests. Comparison of animal data from different groups was analyzed with unpaired t tests. General status and pathological results were described in the report.
Results and Discussion
Construction of pheromonicin-COVID-19
The 28 amino acid sequence of respective antibody mimetic was constructed to follow position I626 of colicin Ia by plasmid synthesis using a pET-11a plasmid containing the colicin Ia gene to form respective Pheromonicin-COVID-19-E (PMC-E) or Pheromonicin- COVID-19-M (PMC-M) or Pheromonicin-COVID-19-M/E (PMCM/ E) or Pheromonicin-COVID-19-E/M (PMC-E/M) (Figure 1A).
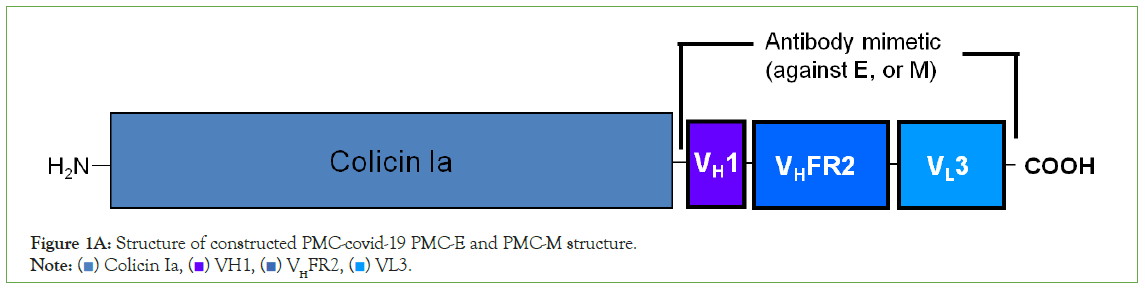
Figure 1A: Structure of constructed PMC-covid-19 PMC-E and PMC-M structure.
Note: ( ) Colicin Ia, (
) Colicin Ia, ( ) VH1, (
) VH1, ( ) VHFR2, (
) VHFR2, ( ) VL3.
) VL3.
Harvested plasmid was transfected into pET B-834 E. coli cells to produce respective PMC-COVID 19s. B834 cell harboring pheromonicin plasmid were grown in LB medium containing 100 μg/ml ampicillin and resuspended in borate buffer. The cells were fractured and debris removed by centrifugation. Nucleic acids were removed by addition of streptomycin sulfate. Dialyzed extracts were applied to a CM-sepharose column. Proteins were recovered by elution with 0.3 M NaCl in borate buffer and collected. The total protein concentration of eluate was about 5-12 mg/ml.
In vitro activity against virus
In vitro neutralization testing was performed to evaluate potential efficacy of PMC-COVID-19 against SARS-CoV-2 infection. Isolated SARS-CoV-2 epidemic (SARS-CoV-2/human/ CHN/CN1/2020), Omicron (BA.1 CCPM-13-V-049-2112-18) and (BA.5 CCPMB- V-049-2207-28) strains were used for viral load inhibition test. After 2-4 hours incubation with PMC-E or PMC-M, the epidemic strain virus titer was reduced 1.5 log10 CCID50/ml lower than that of control. With 3-day daily dosing PMC-E or PMC-M in the virus/ Vero cell culture, the Omicron BA.1 virus copies were reduced about 0.93 log10 copies/ml compared to that of control (Figure 1B). With 2-day daily dosing PMC-E treatment in the virus/Vero cell culture, the Omicron BA.1 virus copies were reduced about 0.55 log10 copies/ml compared to that of control at 48-hours while the virus copies were reduced about 0.25 log10 copies/ml with Nelfinavir/Ritonavir (Pfizer) treatment as a positive control. With 4-day dosing PMC-E/PMC-M or Nelfinavir/Ritonavir treatments in the virus/Vero cell culture, the Omicron BA.1 virus copies were reduced about 0.25 log10 copies/ml compared to that of control (control=about 10.3 log10 copies/ml, PMC, or Nelfinavir/Ritonavir-treated=about 10.05 log10 copies/ml) (Figure 1C). With similar PMC-treatment of BA.5 virus/Vero cell culture, interestingly, the virus load (TCID50/ml) gradually decreased from 3 hours to 72 hours after treatment while the virus load (TCID50/ ml) increased in the Nelfinavir/Ritonavir (Pfizer) positive control (Figure 1D).
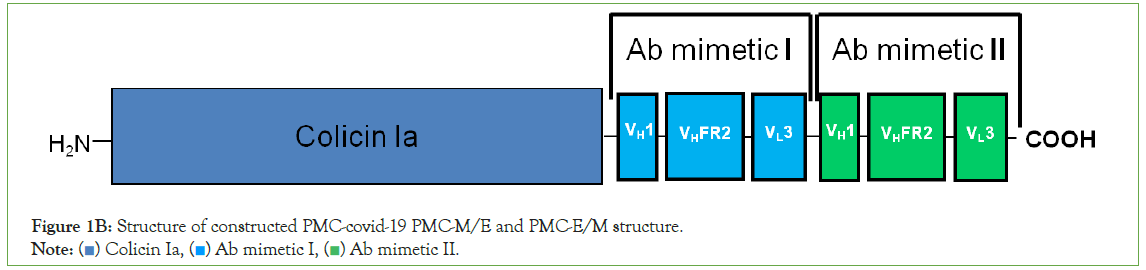
Figure 1B: Structure of constructed PMC-covid-19 PMC-M/E and PMC-E/M structure.
Note: (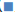 ) Colicin Ia, (
) Colicin Ia, ( ) Ab mimetic I, (
) Ab mimetic I, ( ) Ab mimetic II.
) Ab mimetic II.
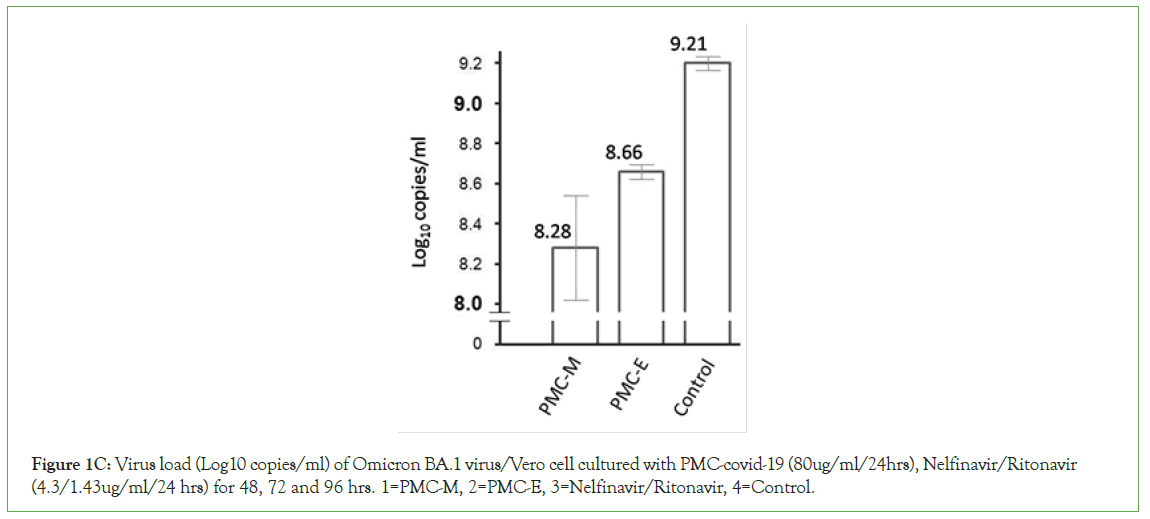
Figure 1C: Virus load (Log10 copies/ml) of Omicron BA.1 virus/Vero cell cultured with PMC-covid-19 (80ug/ml/24hrs), Nelfinavir/Ritonavir (4.3/1.43ug/ml/24 hrs) for 48, 72 and 96 hrs. 1=PMC-M, 2=PMC-E, 3=Nelfinavir/Ritonavir, 4=Control.
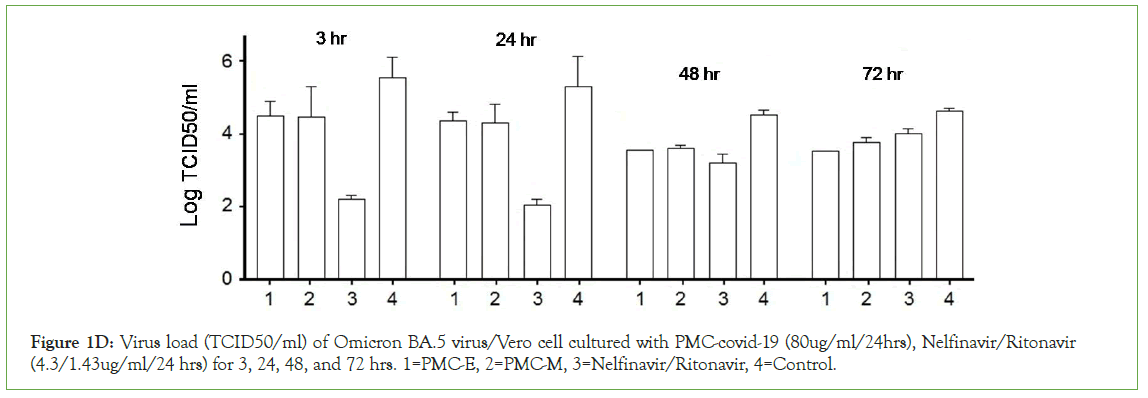
Figure 1D: Virus load (TCID50/ml) of Omicron BA.5 virus/Vero cell cultured with PMC-covid-19 (80ug/ml/24hrs), Nelfinavir/Ritonavir (4.3/1.43ug/ml/24 hrs) for 3, 24, 48, and 72 hrs. 1=PMC-E, 2=PMC-M, 3=Nelfinavir/Ritonavir, 4=Control.
These results indicated that PMC-E and PMC-M were able to inhibit or kill SARS-CoV-2 in vitro and demonstrated broad- spectrum inhibition against tested variants. The PMC-COVID-19 inhibition activity gradually increased while the nirmatrelvir/ ritonavir decreased during the incubation period (Figure 1D). These results suggest that in vivo, the virus/viral-infected eukaryotic cell-killing activity of PMC-COVID-19 might eventually increase with viral-induced immunity to resolve infection.
In vivo activity in coronavirus hamster model
Two tests were performed to evaluate protective efficacy of PMCCOVID- 19 against SARS-CoV-2 respiratory infection. The first test was used to evaluate the PMC-COVID-19 protective efficacy with q24h intraperitoneal injection (IP, 40 μg per gram body weight, 5-day therapy) in 90 male Syrian hamsters, randomly divided into three separate strain/groups of 30 hamsters each were inoculated with three respective variants of SARS-CoV-2, in addition to controls, the PMC-COVID-19 treatment groups were PMC-E, PMC-M, PMC-E/M and PMC-M/E. The second test was used to compare the PMC-COVID-19 protective efficacy with that of neutralizing antibodies in animals inoculated with Delta strain of SARS-CoV-2, the application of PMC-E (n=6) was the same as first test while the neutralized antibodies were applied with q24h intraperitoneal injection (IP, 0.8 mg per animal, 5-day therapy). The Animals were euthanized for lung histopathology and virus load measurement at day 6 post infection.
Hamsters were inoculated with 1 × 105 SARS-CoV-2 virus through nasal drip with SARS-CoV-2 prototype strain (GD108), or South Africa strain (Beta strain, GDPCC-nCpV84), or India strain (Delta strain, CQ79, CSTR,16698.06) respectively. Each strain was inoculated into 5 groups of 6 hamsters each, or 4 groups of 6 hamsters each in the second test. As expected, all controls showed severe interstitial pneumonia with infection. In contrast, all PMC-treated animals were largely protected against SARS-CoV-2 respiratory infection with relatively mild and focal histopathological changes in the lung (Figure 1E-1F). The results of immunolabeling assay demonstrated the distribution of invaded COVID-19 viruses and PMC-COVID-19 molecules which recognized SARS-CoV-2 E proteins and killed viruses/infected host lung cells in the lungs of infected hamsters (Figure 1F).
Figure 1E: The whole lung section of different PMC-E treatments, Delta strain infection, (a) q24h PMC-E treated, 2 animals, (b) control of q24h treatment, 2 animals.
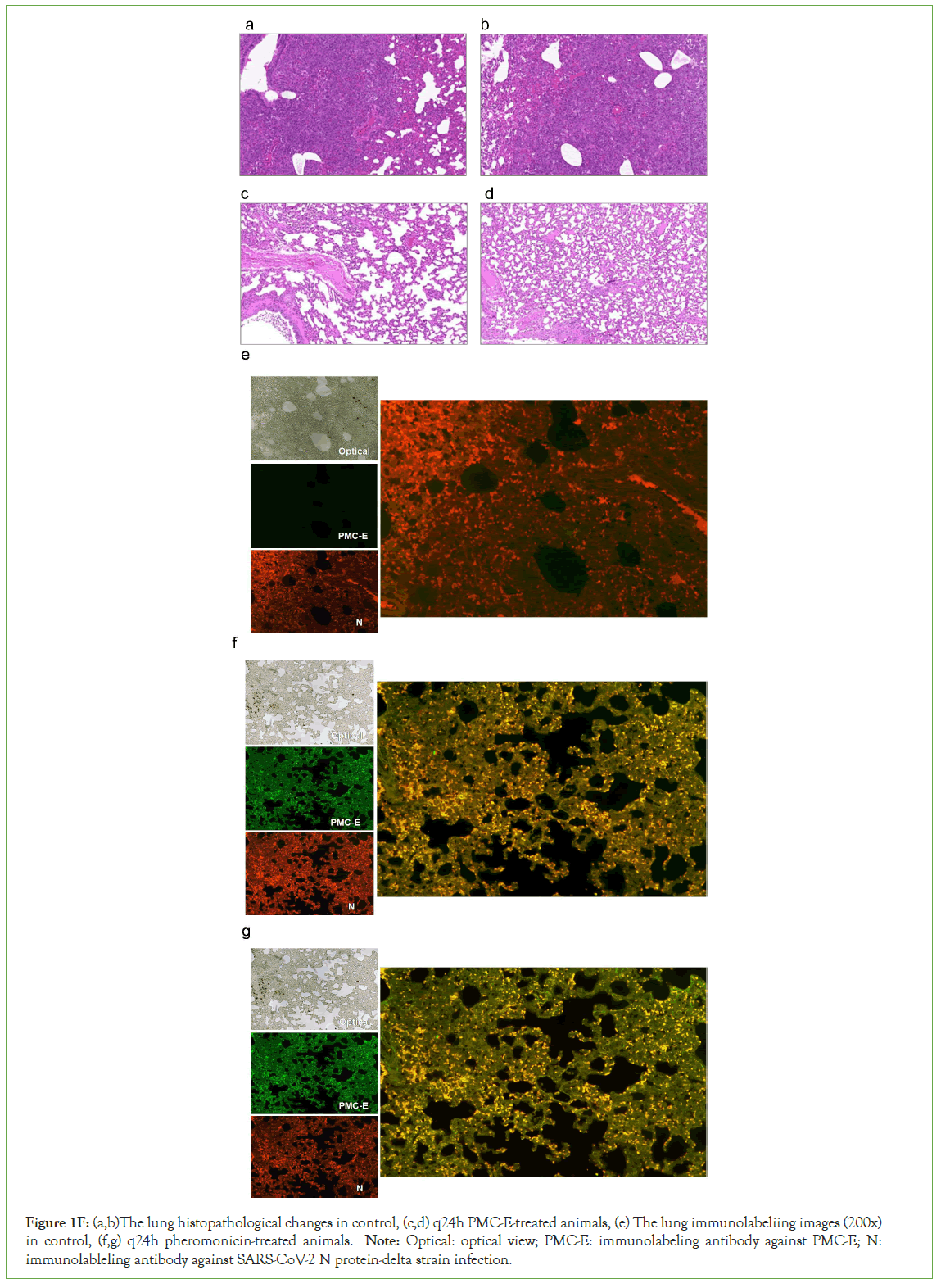
Figure 1F: (a,b)The lung histopathological changes in control, (c,d) q24h PMC-E-treated animals, (e) The lung immunolabeliing images (200x) in control, (f,g) q24h pheromonicin-treated animals. Note: Optical: optical view; PMC-E: immunolabeling antibody against PMC-E; N: immunolableling antibody against SARS-CoV-2 N protein-delta strain infection.
The differences of pulmonary inflammation, structural lesion and hemorrhage between the control and PMC-treatment groups were compared with focal histopathological graded scoring analysis [14]. The results showed that in the Delta strain infected models, the PMC-E (p<0.0001), PMC-M (p<0.01), PMC-E/M (p<0.001) and PMC-M/E (p<0.001) treatments demonstrated significant protective efficacy against viral-induced pulmonary lesions. In the prototype strain infected models, the PMC-E (p<0.0001), PMC-M (p<0.01), PMC-E/M (p<0.0001) and PMC-M/E (p<0.0001) treatments presented significant protective efficacy against viral- induced pulmonary lesions. In the Beta strain infected models, the PMC-E (p<0.01), and PMC-M/E (p<0.05) treatments provided significant efficacy against viral-induced pulmonary lesions (Figure 2A). The PMC-E treatment provided better protective efficacy than neutralizing antibodies against Delta variant-infected models (p<0.05-0.01). Presumably such differences were due to Beta viruses- induced antibodies inability to effectively neutralize the mutated Delta S proteins for protecting viral-induced pulmonary lesions (Figure 2B). These results demonstrated that PMC-COVID-19 provided broad-spectrum in vivo inhibition ability and significant protective efficacy against viral-induced pulmonary lesions.
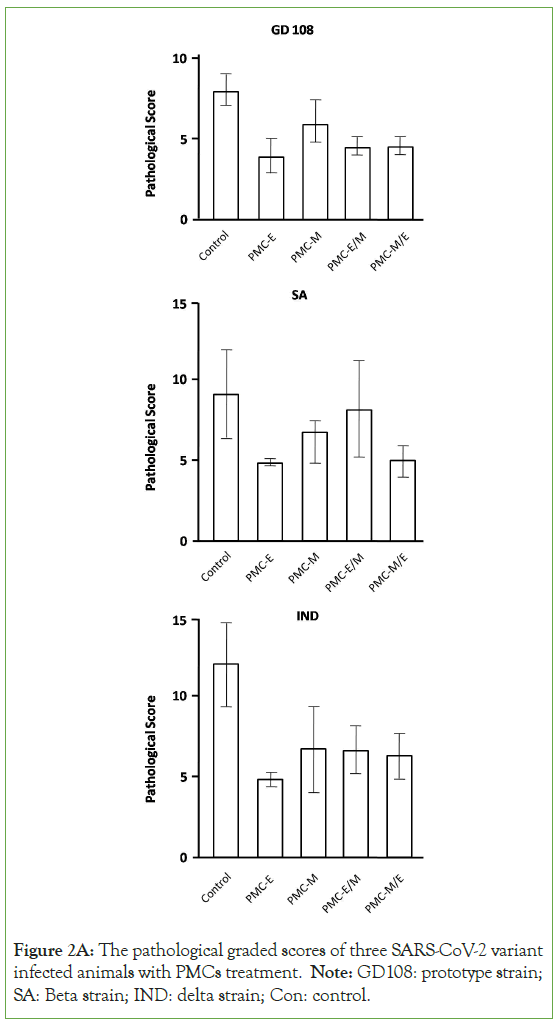
Figure 2A: The pathological graded scores of three SARS-CoV-2 variant infected animals with PMCs treatment. Note: GD108: prototype strain; SA: Beta strain; IND: delta strain; Con: control.
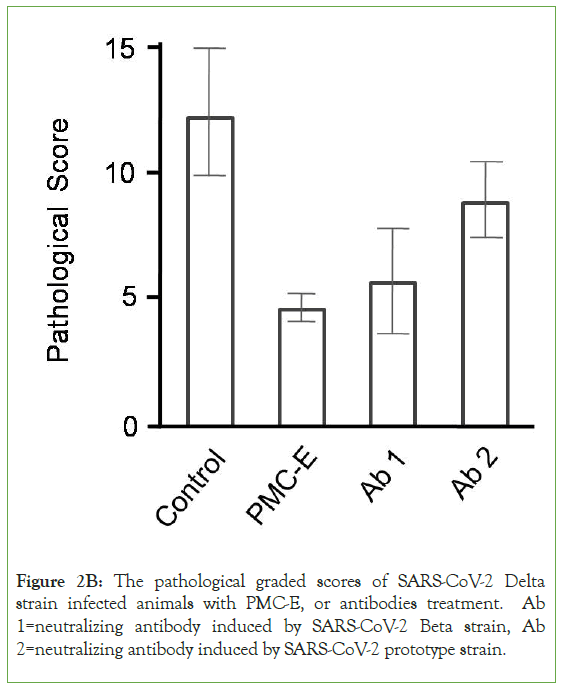
Figure 2B: The pathological graded scores of SARS-CoV-2 Delta strain infected animals with PMC-E, or antibodies treatment. Ab 1=neutralizing antibody induced by SARS-CoV-2 Beta strain, Ab 2=neutralizing antibody induced by SARS-CoV-2 prototype strain.
SARS-CoV-2 infection induced severe inflammatory cytokine storm in the control animals. In contrast, as we found in previous studies, inflammatory cytokines and chemokines appeared at significantly lower levels in pheromonicin-treated animals than that of controls [15]. Above results indicated that pheromonicin might alleviate SARS-CoV-2-induced inflammatory cytokine storm. Indeed it is a vital bioactivity of pheromonicin to save the infected animals from fatal SARS-CoV-2 infection.
The most insurmountable features of SARS-CoV-2 were extensive variation and prominent immunologic escape due to the highly mutated S protein. Interestingly, two other surface proteins, E and M are relatively conserved in present SARS-CoV-2 variants; therefore, there should be little potential for development of resistance. Antigen-antibody neutralization activity could readily yield high occurrence of epitope mutation, however, the antibody mimetic might yield lower occurrence of epitope mutation due to it only recognizing the antigen with lack of subsequent neutralizing activity. It is for this reason that these two proteins were selected as the targets of PMC-COVID-19s, not the S protein. Our in vitro/in vivo results demonstrated that PMC-COVID-19s presented broad- spectrum protective efficacy against tested SARS-CoV-2 variants due to selected targets of PMC-COVID-19s. The broad-spectrum activity against various SARS-CoV-2 variants and the low likelihood of mutations in the E and M proteins suggest that PMC-COVID-19 will be effective against future SARS-CoV-2 variants. The observed PMC-COVID-19 protective efficacy suggests that it could be effective in decreasing fatal SARS-CoV-2 respiratory infections.
The fact that SARS-CoV-2-infected cells are lysed by colicin Ia in PMC-COVID-19 raises significant concerns, in terms of toxicity at higher multiplicity of infections. COVI In-vivo, COVI occurrence of such toxicity would be determined by two factors, (a) native levels of host susceptibility and immunity, there were always around 5%-10% of infected cases would be involved in such toxicity with high susceptibility and hypersensitive immunity; (b) viral, or pheromonicin lysed host cell levels. Previous studies could be considered as a predictor that intracellular pathogen- infected cells lysed with PMC-treatment (PMC-NM, with different 28-residue antibody mimetic against TB) might not raise above significant concerns. Sixteen macaques were infected with high virulence MDR-TB isolate, pulmonary infection was confirmed by tuberculin testing/CT scans 3-wks after inoculation, allocated into three 22-wk treatment groups: untreated controls (n=4), combined INH/RIF-treated (n=4), and anti-TB PMC-treated (n=8, 3 mg/ kg/d). 75% of PMC-treated group survived. All untreated and INH/RIF-treated controls and 25% of PMC-treated group reached the humane endpoint within 10-17 weeks post-infection due to acute tuberculosis pneumonia (Figure 2C). Above results indicated that comparing with untreated control and conventional drug- treated, the lyses of intracellular pathogen infected cells did not present toxicity but prevented further fatal damages from higher multiplicity of infections. In addition, no toxicity of SARS-CoV-2- infected cell lyses was observed in our hamster model (n=72) with 6-day PMC treatment. Further investigation should provide more detailed evidence to alleviate possible toxicity concerns in future clinical trials with PMC-COVID-19. On the other hand, due to the time-lag between the instant virus-cell fusion and delayed E/M protein presentation on infected pulmonary cell membrane, the lysed cell amount of virus-induced destruction was inevitably larger than that of PMC-induced cell destruction (Figure 2D), therefore, "the toxicity at higher multiplicity of infections" should more likely be determined by the virus-lysed cell destruction level, rather than by the infected cell-PMC interaction.
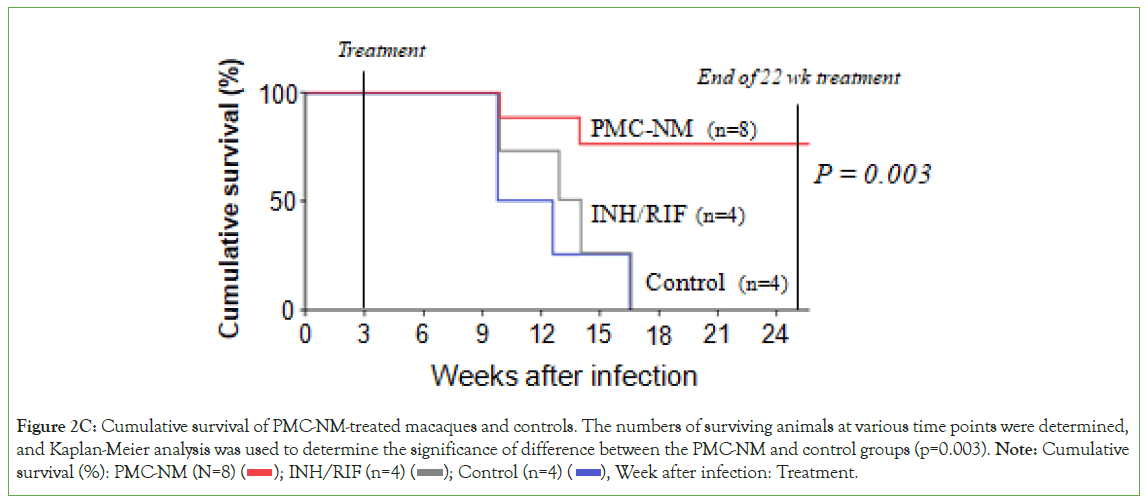
Figure 2C: Cumulative survival of PMC-NM-treated macaques and controls. The numbers of surviving animals at various time points were determined,
and Kaplan-Meier analysis was used to determine the significance of difference between the PMC-NM and control groups (p=0.003). Note: Cumulative
survival (%): PMC-NM (N=8) ( ); INH/RIF (n=4) (
); INH/RIF (n=4) ( ); Control (n=4) (
); Control (n=4) ( ), Week after infection: Treatment.
), Week after infection: Treatment.
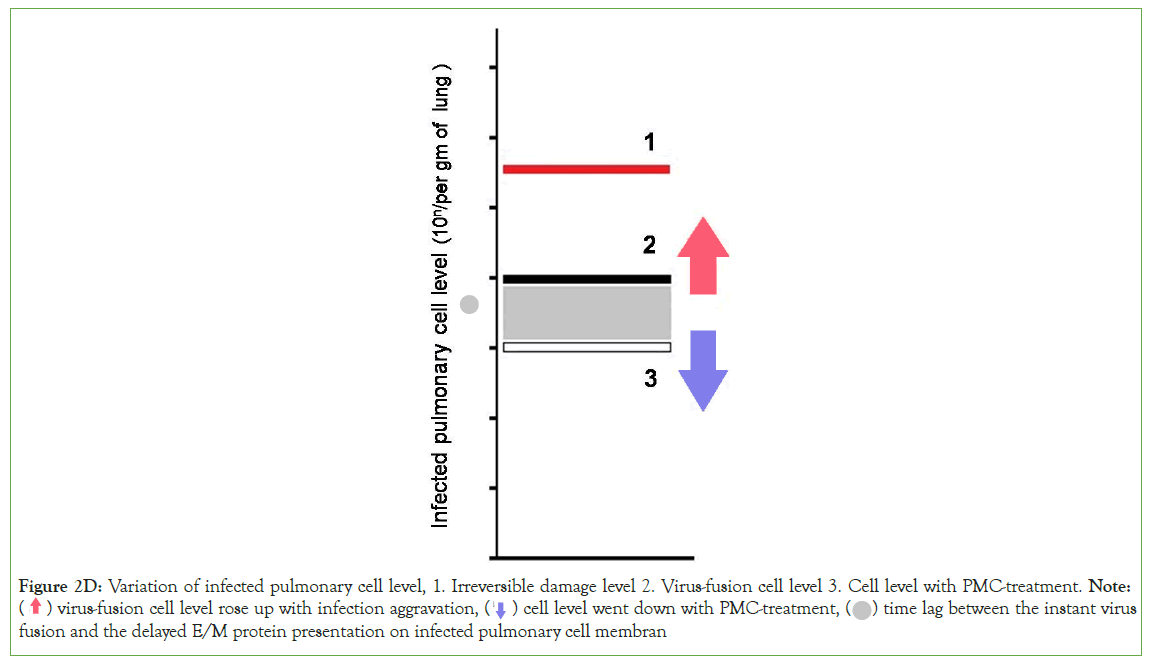
Figure 2D: Variation of infected pulmonary cell level, 1. Irreversible damage level 2. Virus-fusion cell level 3. Cell level with PMC-treatment. Note: ( ) virus-fusion cell level rose up with infection aggravation, (
) virus-fusion cell level rose up with infection aggravation, (  ) cell level went down with PMC-treatment, (
) cell level went down with PMC-treatment, ( ) time lag between the instant virus
fusion and the delayed E/M protein presentation on infected pulmonary cell membran
) time lag between the instant virus
fusion and the delayed E/M protein presentation on infected pulmonary cell membran
Conclusion
Our results indicate that pheromonicin might be an effective platform against SARS-CoV-2 variants. In our assumption, two potential therapeutic roles for PMC-COVID-19 would have a significant impact against the SARS-CoV-2 pandemic, (a) preventive application to close contracts of coronavirus infected cases, (b) early treatments against coronavirus-infected cases with aerosolized inhalation, throat spray, or nasal-spray therapeutics. PMC-COVID-19 requires further evaluation in human clinical trials to define whether it would have a useful role in this setting.
Acknowledgement
This work was supported by National Science and Technology Major Projects of New Drugs (2012ZX09103301 024 and 2015ZX09102007-014), National High Technology Research and Development Program of China (2011AA10A214) of Ministry of Science and Technology; Beijing Municipal Science and Technology Commission (Z131100002713010 and Z161100000116016) to X-Q.Q.
Authors’ Contributions
X-Q.Q, S-Y.Lu, K-F.C, C-Y.Y, Y-N.Z, L-L, R, J-Y.T, D.Z, F-Y.L, H-F.L, R-Q.L, and X-Z. P prepared antibody mimetics and pheromonicin- COVID-19 molecules, measured in vitro and in vivo activity; X-Q.Q. designed and organized the study and manuscript. Q iu XQ and Lu SY both authors equally contributed.
Competing Interests
All authors declare that they have no conflicts of interest.
References
- Wang Y, Zhang D, Du G, Du R, Zhao J, Jin Y, et al. Remdesivir in adults with severe COVID-19: A randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395(10236):1569-1578.
- Sohrabi C, Alsafi Z, O'neill N, Khan M, Kerwan A, Al-Jabir A, et al. World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). Int J Surg. 2020;76:71-76.
- Kannan S, Shaik Syed AP, Sheeza A. Omicron (B. 1.1. 529)-variant of concern-molecular profile and epidemiology: A mini review. Eur Rev Med Pharmacol Sci. 2021;25(24):8019-8022.
- Cao Y, Wang J, Jian F, Xiao T, Song W, Yisimayi A, et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602(7898):657-663.
- Koley T, Kumar M, Goswami A, Ethayathulla AS, Hariprasad G. Structural modeling of Omicron spike protein and its complex with human ACE-2 receptor: Molecular basis for high transmissibility of the virus. Biochem Biophys Res Commun. 2022;592:51-53.
- Hoffmann M, Krüger N, Schulz S, Cossmann A, Rocha C, Kempf A, et al. The Omicron variant is highly resistant against antibody-mediated neutralization: Implications for control of the COVID-19 pandemic. Cell. 2022; 185(3):447-456.
- Wang Z, Yang L. Broadâ?spectrum prodrugs with antiâ?SARSâ?CoVâ?2 activities: Strategies, benefits, and challenges. J Med Virol. 2022; 94(4):1373-1390.
- Yao TT, Qian JD, Zhu WY, Wang Y, Wang GQ. A systematic review of lopinavir therapy for SARS coronavirus and MERS coronavirus-A possible reference for coronavirus diseaseâ?19 treatment option. J Med Virol. 2020; 92(6):556-563.
- Qiu XQ, Riley MA. In The Bacteriocins: current knowledge and future prospects, Dorit R, Roy SM. & Riley MA, Eds. Capturing the Power of Van der Waals Zone in the Creation of a Novel Family of Bacteriocinbased Antibiotics. 2016;4: 65-80.
[Crossref]
- Qiu XQ, Wang H, Lu XF, Zhang J, Li SF, Cheng G, et al. An engineered multidomain bactericidal peptide as a model for targeted antibiotics against specific bacteria. Nat Biotechnol. 2003;21(12):1480-1485.
- Qiu XQ, Zhang J, Wang H, Wu GY. A novel engineered peptide, a narrow-spectrum antibiotic, is effective against vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother. 2005;49(3):1184-1189.
- Qiu XQ, Tong CY, Zhong ZQ, Wang WQ, Zuo YW, Huang Y, et al. An engineered multidomain fungicidal peptide against plant fungal pathogens. Act Physiol Sin. 2013;65(4):417-432.
- Qiu XQ, Wang H, Cai B, Wang LL, Yue ST. Small antibody mimetics comprising two complementarity-determining regions and a framework region for tumor targeting. Nat Biotechnol. 2007;25(8):921-929.
- Liu L, Wei Q, Lin Q, Fang J, Wang H, Kwok H, et al. Antiâ??spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4(4).
- Qiu XQ, Cao KF, Zhang XF, Tong CY, Ma HL, Xu HM, et al. Defending the homeland: Microbiome molecules provide protection to their vertebrate hosts. Future Microbiol. 2020;15(18):1697-1712.
Citation: Qiu XQ, Lu SY, Cao KF, Tang JY, Zhang D, Luo FY, et al. (2023) A Broad-Spectrum Antiviral Fusion Protein, Pheromonicin Demonstrated Protective Efficacy Against SARS-CoV-2 Variants in vitro and in vivo Models. Clin Microbiol. 12: 328.
Copyright: © 2023 Qiu XQ, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
