Indexed In
- Open J Gate
- Genamics JournalSeek
- CiteFactor
- RefSeek
- Hamdard University
- EBSCO A-Z
- NSD - Norwegian Centre for Research Data
- OCLC- WorldCat
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2016) Volume 5, Issue 1
Two Novel Somatic Mutations in Exon 15 of the Adenomatous Polyposis Coli Gene in Iranian Familial Adenomatousis Polyposis Coli Patients
Abstract
Background: The Adenomatous Polyposis Coli (APC) gene is considered as a gatekeeper in colorectal tumorigenesis. 60% of somatic mutations in the APC gene are concentrated in a region called the Mutation Cluster Region (MCR). In this study, our objective was to perform the genetic analysis of two patients (index patients) who had been selected by colorectal cancer features, and to identify the genetic changes in the MCR region of the APC gene. Following discussions about the disease, the patients (index persons) agreed to a further genetic evaluation.
Materials and methods: Mutation analysis of the MCR, which spans codons 1286-1513, was performed on the paraffin-embedded cancerous tissue samples using macro dissection, nested PCR and direct sequencing of purified PCR fragments.
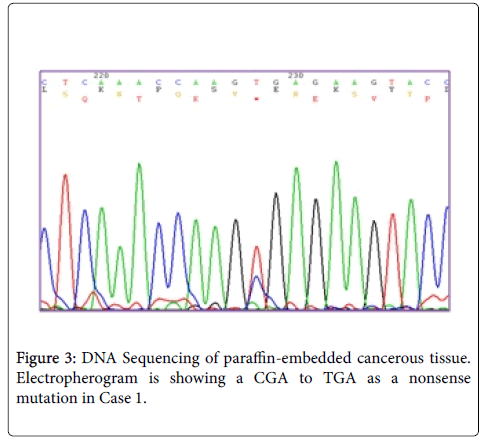
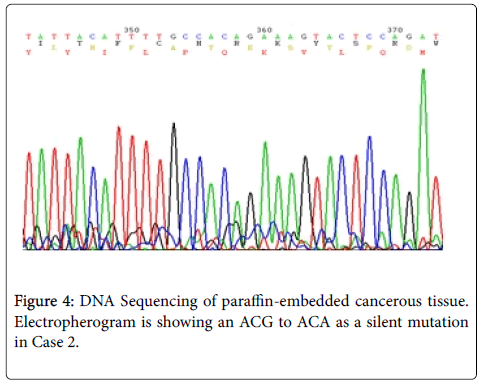
Results: In our study, two new somatic mutations detected in these patients. In one patient, we have detected a CGA to TGA as a Nonsense mutation that lead to Arg to premature Stop codon at the 4507 nucleotide position (Codon 1503) and in another patient, we describe a G→A Transition (ACG to ACA) at nucleotide position 4638 in exon 15 (MCR) which causes a silent mutation since both normal and mutated alleles encode a Thr residue at codon 1546. These mutations have not been described previously.
Conclusion: This observation could suggest differences in the frequency of pathological mutations in APC among different populations; however, epidemiological studies must be performed to confirm this theory which it is not the aim of our present work.
Keywords: Adenomatous polyposis coli, Iranian familial adenomatousis polyposis, Somatic mutations
Introduction
Familial Adenomatous Polyposis (FAP) is a rare autosomal dominant genetic disease, with an approximate incidence rate of 1/8000 [1]. Clinical manifestations are mainly multiple adenomatous polyps in the large intestine (more than 100 polyps in total), and most of these patients fall ill in adolescence. Adenomatous polyps are a type of precancerous lesions. Thereby, cancerization will occur before the age of 40 years in almost 100% of patients without treatment. It was shown the occurrence of FAP was related to adenomatous polyposis coli gene mutations found at 5q21-q22 [2]. APC gene micro mutations were identified in about 60-70% of FAP patients [3], whereas large fragment deletion mutations of APC gene were identified in 10-15% [3,4]. It is also helpful for monitoring and in the clinical treatment of high-risk individuals with mutant genes and can effectively decrease the incidence and mortality of FAP [5]. Based on the model provided by the autosomal dominant condition, familial adenomatosis polyposis, it has long been assumed that APC mutation is the starting alteration is most colorectal cancers [6]. APC mutation is more frequent in rectal than colonic cancer [7]. The APC gene includes 21 exons contained within a 98-kilobase locus [8]. The largest exon, 15, comprises more than 75% of the 8,535 base pairs (bp) of the coding sequence. And is the target of most germline mutations in FAP patients and somatic mutations in tumors [9-11]. 60% of somatic mutations in the APC gene are concentrated in a region called the Mutation Cluster Region (MCR) [12]. Somatic mutations are clustered in the central region of the open reading frame, spanning codons 1286-1513 of exon 15 [13,14]. Although this MCR represents only 8% of the 8535 bp APC gene, 66-77% of somatic mutations in APC occur in this region [15]. The APC protein consists of 2843 amino acid and comprises several functional domains, motifs and other signatures, as reviewed by polakis [14].
Materials and Methods
Patient’s clinical features
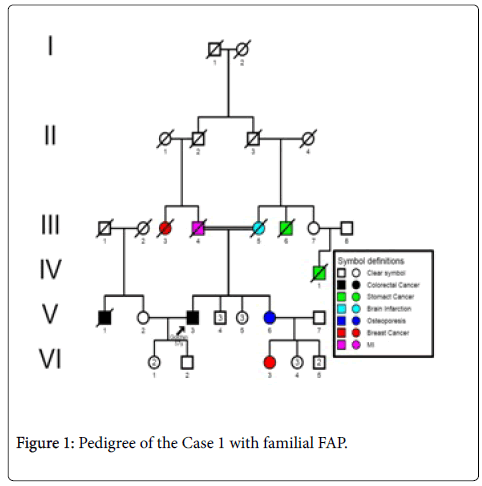
Case 1: A 57 years- old male patient (index person) with a familial history of FAP with onset age at 53 years, resulting of a consanguinity marriage (Figure 1) with recurrent haematochezia. Colonoscopy was performed and friable, bleeding lesion and 5 polypi was found at the lineal flexure of the large bowel with circular narrowing of the lumen. The histopathological report of the biopsy showed a mucinous adenocarcinoma infiltrating the muscularis properia. Ultrasound ruled out evidence of liver metastasis.
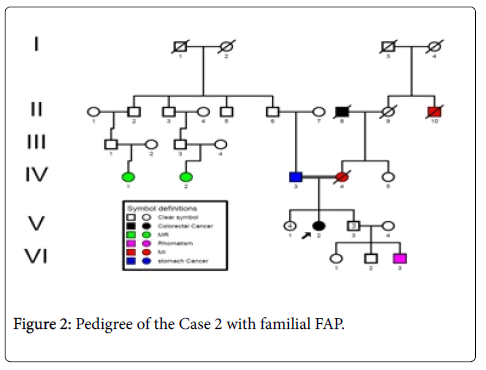
Case 2: A 60 years- old male patient (index person) with a family history of FAP (Figure 2), resulting of a non-consanguinity marriage and with onset age of 56 years, with anemia and Iron deficiency was detected in the laboratory testing. Colonoscopy was performed and more than 10 adenomatous polyps have been seen. Ultrasound ruled out evidence of liver metastasis.
DNA extraction
After obtaining the Consent form from the patient, DNA was extracted from macrodissected paraffin-embedded cancerous tissue sample of the index patients following deparaffinization and protein K digestion, according to the protocol of the high pure PCR Template purification kit (Roche Diagnostics GmbH, Mannheim, Germany). DNA concentration and purity was measured in a spectrophotometer at 260 and 280 nm.
APC mutation analysis
Since most of the somatic mutations in APC occur within the MCR, We amplified the MCR as four overlapping fragments (codons 1286-1520) in a nested PCR strategy. Flank PCR was performed to generate two fragments A and B. Genomic DNA (250 ng) was added to 50 Mm KCL, 10 mM Tris-HCL (PH 8.3), 2.0 Mm Mgcl2, 250 mM deoxyribonucleotide triphosphate (dNTP), (pharmacia, Uppsala, Sweden), 200 nM of each primer (Table 1) [15], and 1 U platinum Taq (Invitrogen, B1eda, The Netherlands) in a final volume of 50 μl. Fragment A was used as starting material for amplifying nested fragments S1 and S2, and fragment B was used for nested fragments S3 and S4. An alternative semi- nested PCR strategy was chosen when nested PCR amplification failed for one or more fragments originating from flank A or B. Alternative flank fragments F1 to F4 were then generated for each of the four nested fragments S1 to S4, respectively. The semi-nested PCR was performed in a final volume of 50 μl, containing 5 μl of a 1:100 dilution of the flank products, 50 mM KCL, 10 Mm Tris-HCL(PH 8.3), 1.5-2.5 mM MgCl2, 250 mM dNTP, 200 nM of each primer (Table 1) [15] and 1 U platinum Taq. The cycles for both flank, nested and semi-nested PCR consisted of 3 min predenaturation at 94°C, 40 s at 94°C for denaturation, 1 min at 52°C to 57°C for annealing, 1 min at 72°C for elongation and 10 min at 72°C for post-elongation. The length and concentration of the PCR products were checked by electrophoresis on 2% agarose gels and visualized with ethidium bromide. Mutation analysis was performed by an ABI 3130 automated sequencer (XL Genetic Analyser) using the Big-Dye Terminator Version 3.1 Cycle Sequencing method.
| Case Number | Gene/Exon | Nucleotide Position | Codon/Mutation | Amino acid | Type |
|---|---|---|---|---|---|
| 1 | APC(MCR)/15 | 4507 | 1503 (CGA → TGA) | Arg → Stop codon | Nonsense |
| 2 | APC(MCR)/15 | 4638 | 1546 (ACG → ACA) | Th r→ Thr | Silent |
Table 1: Type and location of detected mutation
Results and Discussions
Sequence analyzes were performed by Bio Edit software. For the changed base sequence, the mutational site and type were determined in National Center for Biotechnology Information, and then the mutations were identified as new by referring to the mutations reported in the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/gene.php.gene=APC) and the UMD-APC mutations database (http://www.umd.be/APC/). We studied two Iranian patients with the typical clinical symptoms of FAP. A survey of the two new somatic mutations detected in these patients is summarized in Table 2. In one patient (case 1) we have detected a CGA to TGA as a Nonsense mutation that lead to Arg to premature Stop codon at the 4507 nucleotide position (Codon 1503) (Figure 3) and in another patient (case 2), we describe a G→A transition at nucleotide position 4638 in exon 15 (MCR) which causes a silent mutation since both normal and mutated alleles encode a Thr residue at codon 1546 (Figure 4). This mutation has not been described previously. The large number of APC mutations have been reported so far, and most of the mutations lead to truncation of the APC protein either by a nonsense mutation (30%) or by a frameshift mutation (68%) and most of the mutations (germline or somatic) occurred in the first half of the coding region. In somatic mutations, two hot spots occur at positions 1309 and 1450 that this concentration of mutations in the 5′part of the gene that leading to the synthesis of truncated APC [16] whereas in our study a new mutation is found at position 1503 in an FAP patient with typical clinical symptoms. Hence, this observation could suggest differences in the frequency of pathological mutations in APC among different populations; however, epidemiological studies must be performed to confirm this theory which it is not the aim of our present work.
References
- Giardiello FM, Krush AJ, Petersen GM, Booker SV, Kerr M, et al. (1994) Phenotypic variability of familial adenomatous polyposis in 11 unrelated families with identical APC gene mutation. Gastroenterology 106: 1542-1547.
- Leppert M, Dobbs M, Scambler P, O'Connell P, Nakamura Y, et al. (1987) The gene for familial polyposis coli maps to the long arm of chromosome 5. Science 238: 1411-1413.
- Michils G, Tejpar S, Thoelen R, van Cutsem E, Vermeesch JR, et al. (2005) Large deletions of the APC gene in 15% of mutation-negative patients with classical polyposis (FAP): a Belgian study. Hum Mutat 25: 125-134.
- Nieuwenhuis MH, Vasen HF (2007) Correlations between mutation site in APC and phenotype of familial adenomatous polyposis (FAP): a review of the literature. Crit Rev Oncol Hematol 61: 153-161.
- Vasen HF, Möslein G, Alonso A, Aretz S, Bernstein I, et al. (2008) Guidelines for the clinical management of familial adenomatous polyposis (FAP). Gut 57: 704-713.
- Fodde R, Smits R, Clevers H (2001) APC, signal transduction and genetic instability in colorectal cancer. Nat Rev Cancer 1: 55-67.
- Kapiteijn E, Liefers GJ, Los LC, Kranenbarg EK, Hermans J, et al. (2001) Mechanisms of oncogenesis in colon versus rectal cancer. J Pathol 195: 171-178.
- Thliveris A, Albertsen H, Tuohy T, Carlson M, Groden J, et al. (1996) Long-range physical map and deletion characterization of the 1100-kb NotI restriction fragment harboring the APC gene. Genomics 34: 268-270.
- Thliveris A, Samowitz W, Matsunami N, Groden J, White R (1994) Demonstration of promoter activity and alternative splicing in the region 5' to exon 1 of the APC gene. Cancer Res 54: 2991-2995.
- Horii A, Nakatsuru S, Ichii S, Nagase H, Nakamura Y (1993) Multiple forms of the APC gene transcripts and their tissue-specific expression. Hum Mol Genet 2: 283-287.
- Suleková Z, Ballhausen WG (1995) A novel coding exon of the human adenomatous polyposis coli gene. Hum Genet 96: 469-471.
- Béroud C, Soussi T (1996) APC gene: database of germline and somatic mutations in human tumors and cell lines. Nucleic Acids Res 24: 121-124.
- Nagase H, Nakamura Y (1993) Mutations of the APC (adenomatous polyposis coli) gene. Hum Mutat 2: 425-434.
- Polakis P (1997) The adenomatous polyposis coli (APC) tumor suppressor. Biochim Biophys Acta 1332: F127-F147.
- Lüchtenborg M, Weijenberg MP, Roemen GM, de Bruïne AP, van den Brandt PA, et al. (2004) APC mutations in sporadic colorectal carcinomas from the Netherlands cohort study. Carcinogenesis 25: 1219-1226.
- Su LK, Johnson KA, Smith KJ, Hill DE, Vogelstein B, et al. (1993) Association between wild type and mutant APC gene products. Cancer Res 53: 2728-2731.
Copyright: © 2016 Reza V. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.