Indexed In
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- Ulrich's Periodicals Directory
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
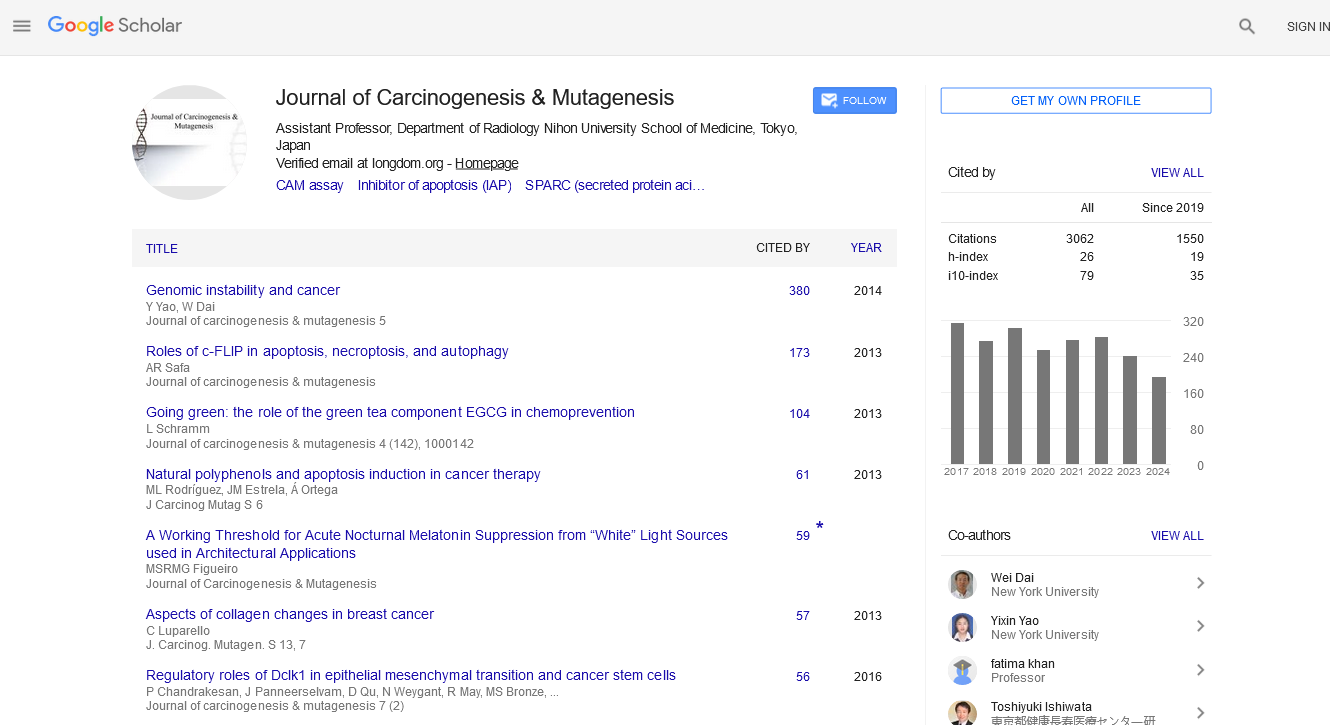
Commentary - (2025) Volume 0, Issue 0
The Role of Whole Genome Sequencing in Understanding Cancer Mutations
Guangjun Wang*Received: 30-Apr-2025, Manuscript No. JCM-25-29376; Editor assigned: 02-May-2025, Pre QC No. JCM-25-29376 (PQ); Reviewed: 16-May-2025, QC No. JCM-25-29376; Revised: 23-May-2025, Manuscript No. JCM-25-29376 (R); Published: 30-May-2025, DOI: 10.35248/2157-2518.25.S50.002
Description
Whole Genome Sequencing (WGS) has emerged as a revolutionary tool in modern biomedical research, offering unprecedented insights into the genetic architecture of diseases, including cancer. By allowing researchers to analyze the complete DNA sequence of an organism's genome at a single time, WGS provides a comprehensive overview of genetic alterations that may underlie carcinogenesis and mutagenesis. This method enables the identification of point mutations, insertions, deletions, copy number variations, and structural rearrangements, making it a powerful approach to elucidate the molecular mechanisms driving tumor initiation, progression, and resistance to therapy.
In cancer research, WGS has been instrumental in cataloging somatic mutations across various tumor types. Large-scale initiatives such as The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC) have utilized WGS to characterize the mutational landscape of numerous malignancies, revealing both common and rare driver mutations. These findings have led to the development of molecular subtypes of cancer, allowing for more precise classification and improved prognostic and therapeutic strategies. For instance, in breast cancer, WGS has helped distinguish between luminal A, luminal B, HER2-enriched, and basal-like subtypes based on distinct mutational and transcriptomic signatures.
Furthermore, WGS facilitates the discovery of novel cancer-associated genes that may not be captured by targeted or exome sequencing approaches. This is particularly important in rare cancers or those with atypical presentations, where the underlying mutations may reside in non-coding regions or in genes not previously associated with malignancy. By identifying such mutations, WGS expands the repertoire of potential biomarkers and therapeutic targets, contributing to the advancement of personalized oncology.
Another critical application of WGS in carcinogenesis research is the detection of mutational signatures, which are patterns of mutations resulting from specific mutagenic processes. These signatures can provide valuable information about the etiological factors contributing to cancer development. For example, ultraviolet radiation, tobacco smoke, and defects in DNA repair mechanisms leave distinct mutational imprints on the genome. By analyzing these signatures, researchers can trace the origins of tumors and better understand the environmental and endogenous factors involved in carcinogenesis.
Whole genome sequencing also plays a pivotal role in understanding tumor heterogeneity and clonal evolution. Cancers are composed of diverse subclones with distinct genetic profiles, which can influence disease progression and treatment response. WGS allows for the reconstruction of the evolutionary history of tumors by identifying clonal and subclonal mutations. This information is crucial for designing effective therapeutic strategies, as it helps in anticipating potential resistance mechanisms and optimizing treatment regimens.
In the realm of mutagenesis, WGS provides a detailed map of genomic instability and mutation burden in cancer cells. It enables the identification of regions prone to genomic rearrangements, such as chromothripsis or kataegis, which are hallmarks of certain cancers. Understanding these patterns of instability can shed light on the underlying defects in DNA repair pathways and inform the development of targeted therapies, such as PARP inhibitors for tumors with BRCA mutations.
In conclusion, whole genome sequencing stands at the forefront of cancer genomics, offering deep insights into the genetic alterations and mutagenic processes driving malignancies. As technology evolves and our understanding of cancer biology deepens, WGS is poised to become an integral part of both research and clinical oncology, ultimately paving the way for more precise, predictive, and personalized approaches to cancer diagnosis and treatment.
Citation: Wang G (2025). The Role of Whole Genome Sequencing in Understanding Cancer Mutations. J Carcinog Mutagen. S50:002.
Copyright: © 2025 Wang G. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited