Indexed In
- Academic Journals Database
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- ResearchBible
- Ulrich's Periodicals Directory
- Electronic Journals Library
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research - (2022) Volume 13, Issue 2
The Complete Mitochondrial Genome Sequence Variation and Phylogenetic Analysis of Mulberry
Guo Liangliang1, Shi Yisu1, Wu Mengmeng1, Michael Ackah1, Qiang Lin2 and Weiguo Zhao1*2Sericultural Research Institute, Guangxi Zhuang Autonomous Region, Nanning,530007, P.R, China
Received: 01-Mar-2022, Manuscript No. JDMGP-22-15600; Editor assigned: 04-Mar-2022, Pre QC No. JDMGP-22-15600 (PQ); Reviewed: 18-Mar-2022, QC No. JDMGP-22-15600; Revised: 25-Mar-2022, Manuscript No. JDMGP-22-15600 (R); Published: 04-Apr-2022, DOI: 10.4172/2153- 0602.22.13.249
Abstract
Background: Mulberry is an economically significant crop, tolerance to various environmental conditions. The plant (leaves) is use for feeding silkworm and, for its landscaping and possesses high development prospects and scientific research value. Mitochondria are the plants' powerhouse that produces the required energy to carry out life processes.
Objective: Plant mitochondria (mt) genome serves as a powerhouse that produces the required energy to carry out life processes in plants. However, the mitochondria (mt) genome of mulberry plant is still unexplored. This study investigated the mt genome of Morus L (M. atropurpurea and M. multicaulis) and compared it to other plant species.
Methods: The mt genome of Morus L (M. atropurpurea and M. multicaulis) were sequenced using Oxford Nanopore Prometh ION and data assembled and analyzed and then compared to other plants mitochondrion genome. Phylogenetic analysis was arried on to study the evolution status of the mulberry plants studied
Results: The circular mt genome of M. multicaulis has a length of 361,546 bp, contains 54 genes, including 31 protein-coding genes, 20 tRNA genes, and 3 rRNA genes and composition of A (27.38%), T (27.20%), C (22.63%) and G (22.79%). On the hand, the circular mt genome of M. atropurpurea has a length of 395,412 bp long, comprises C+G (45.50%), including 57 functional genes containing 2 rRNA genes, 22 tRNA genes and 32 PCGs. There exist sequence repeats, RNA editing gene and migration from cp to mt in the M. multicaulis and M. atropurpurea mt genome.
Phylogenetic analysis based on the complete mt genomes of Morus and other 28 species reflect an exact evolutionary and taxonomic status.
Conclusion: We found out that the Morus species mt genome is circular, with M. multicaulis having a length of 361,546 bp. 54 genes, including 31 protein-coding genes, 20 tRNA genes, and 3 rRNA genes. Also, M. atropurpurea was found to have a length of 395,412 bp. Moreover, a total of 57 genes contains 32 protein-coding genes, 22 tRNA and 3 rRNA were annotated in the genome. The results will provide a comprehensive understanding of the Morus mt genome and may help in future studies and breeding of mulberry varieties.
Keywords
M. multicaulis; M. atropurpurea; Mitochondrial genome; Variation; Phylogenetic analysis.
Introduction
Mulberry plant is a native to China and is an economically significant plant belonging to the Moraceae family. The leaves of the plant are mostly used in China to feed silkworm insect. In terms of environmental protection, the plant is used worldwide for erosion control and windbreaks. Aside from its usage for feeding silkworm, mulberry has other great value as a source of food for a healthy life. In medicine, the plant is used as herbal medicine to cure fever, improve eyesight, strengthen joints, and lower blood pressure in China [1].
Mitochondrial (mt) genome is a power source for energy synthesis and conversion. It provides energy protection for various cells’ physiological activities [2]. These include cell differentiation, apoptosis, cell growth [3]. The mt genome is involved in the synthesis and degradation of several compounds, therefore, it plays an essential role in plant productivity and development [4,5]. The mt genome is highly conserved but varies in length, gene sequence, and content [6]. Most plant mt varies from 200 kb to 3 Mb and more extensive than other eukaryotes’ mt genomes [7]. The smallest known terrestrial plant mt is about 66 kb, and the most extensive terrestrial plant mt genome length is 11.3 Mb [8,9]. The mt genome structures are shaped by active recombination, gene transfer to the nucleus and other forces such as physical mapping and sequencing that remain unclear, contributes to some of the smallest mt genomes [10].
Structural analyses revealed high intra- and intermolecular recombination frequencies, which generated a structurally dynamic assemblage of genome configurations [11]. The mt genome is inherited from the maternal parent; this provides a powerful model for studying genome structure and evolution and certain advantages in phylogenetic reconstruction. These genomes exhibit an intriguing mixture of conservative (slowest rates of nucleotide substitution) and dynamic evolutionary patterns [12]. Previous reports suggest that it is unnecessary for evolutionary studies to assemble whole organelle genomes, but studies should consider exploring the variations [13].
With the rapid development of sequencing technology, an increasing number of complete plants mt genomes have been assembled and reported. Currently, 351 complete mt genomes have been deposited in GenBank Organelle Genome Resources [14]. However, the mt genome of Morus is incomplete and unexplored. In this study, we sequenced and annotated the mt genome of cultivated Morus (M. atropurpurea and M. multicaulis) and then compared them to the wild M. notabilis (NC-041177.1) and other eudicots to investigate the mt genome structure, repeat sequences, phylogenetics and others. The findings of this study will provide additional information for a better understanding of the genetics of the Morus L.
Materials and Methods
Plant material, DNA extraction, and sequencing
The M. atropurpurea and M. multicaulis plants were collected from National Mulberry GenBank Zhenjiang City, Jiangsu Province, China.
Plant Genomic DNA Kit was used to isolating total genomic DNA from 100 mg fresh leaves. DNA sample quality was examined with the Nanodrop instrument and agarose-gel electrophoresis. The quality DNA samples were then sent to (Genepioneer Biotechnology company, Nanjing, China) for sequencing using Oxford Nanopore Prometh ION. The second and third generation sequencing strategies was used in this study.
Quality control of sequencing data
Sequencing using Oxford Nanopore Prometh ION platform was performed on the two mulberry M. multicaulis and M. atropurpurea. The data quality was checked using fastp software (version 0.20.0) at the default parameters. To improve the accuracy of the analysis, the Raw Reads were filtered again according to the following criteria; (i) removal of the sequenced connectors and primer sequences in reads (ii) reads with an average mass value less than Q5 were filtered out (iii) reads with N number greater than 5 were removed. The quality reads after the above checks, called clean reads were subjected to subsequent analysis.
Assembly and annotation of the mitochondrial genome
The mt genome sequence of mulberry was selected using blast v2.6 (https://blast.ncbi.nlm.nih.gov) /Blast.cgi). The contig was aligned with the plant mitochondrial gene database (the mitochondrial gene sequence of the species published on NCBI). They were subsequently assembled by using Canu software with the selected reads. NextPolish1.3.1 (https://github.com/Nextomics/NextPolish) was assigned to calibrate and pilon was used to correct read errors to get the final assembly results. The encoded protein and rRNA were aligned to the published plant mitochondrial sequence as a reference and then further adjustments were made according to the relative species.
The tRNAscanSE (http://lowelab.ucsc.edu/tRNAscan-SE/) was used to annotate the tRNA. The open reading frame (ORF) was annotated using open reading frame finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The minimum length was set to 102 bp to exclude redundant sequences and sequences that overlap with known genes. Sequence alignments longer than 300 were annotated against the nr library. Results were obtained after checking and manually confirmed the final annotation. The circular mitochondrial genome map was drawn using OGDRAW (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html).
Analysis of repeated sequences
The scattered repetitive sequences were detected using vmatch v2.3.0 (http://www.vmatch.de/), combining Perl scripts to identify repetitive sequences with a minimum length set to 30 bp and hamming distance 3.
RNA editing analyses and chloroplast to mitochondrion DNA transformation
The online sites (http://www.prepact.de/prepact-main.php) was used to predict the editing sites in the mitochondrial RNA of M. multicaulis and M. atropurpurea. The cpDNA of M. multicaulis (KU355297) and M. atropurpurea (KU355276) was downloaded from NCBI Organelle Genome Resources Database. We used blast software to set similarity to 70% and e-value to 10E-5 and Circos (v0.69) was used to draw the map for the data visualization.
Variation architecture and phylogenetic tree construction
Nucleic acid diversity (pi) was performed by maft software (set to default) to compare the homologous gene sequences of distinct species globally, and dnasp5 was used to calculate the variation. Comparison of the mt genome sequence with other plastomes at the global level was made using mVISTA online software in shuffle-LAGAN mode. MEGA7.0 software was used to construct the phylogenetic tree by utilizing the Maximum Likelihood (ML) and Neighbor-Joining (NJ) methods with a bootstrap of 1,000 Poisson models. The M. notabilis (NC-041177.1) mt genome data was downloaded from NCBI.
Results
Analysis of sequencing data and quality control
An overview of the mitochondrial sequencing reads derived from the M. multicaulis M. atropurpurea libraries is listed in Table1. A total of 33780517 and 29282471 raw reads were obtained from M. atropurpurea and M. multicaulis, respectively. The sequencing depth of M. multicaulis is169 x and M. atropurpurea is 155 x. After quality control check on the raw reads, 1112754 and 930791 clean reads were obtained from M. atropurpurea and M. multicaulis, respectively (Table 2). In addition, the sequencing read lengths as well as the base distribution in mulberry plants are shown in Figure 1.
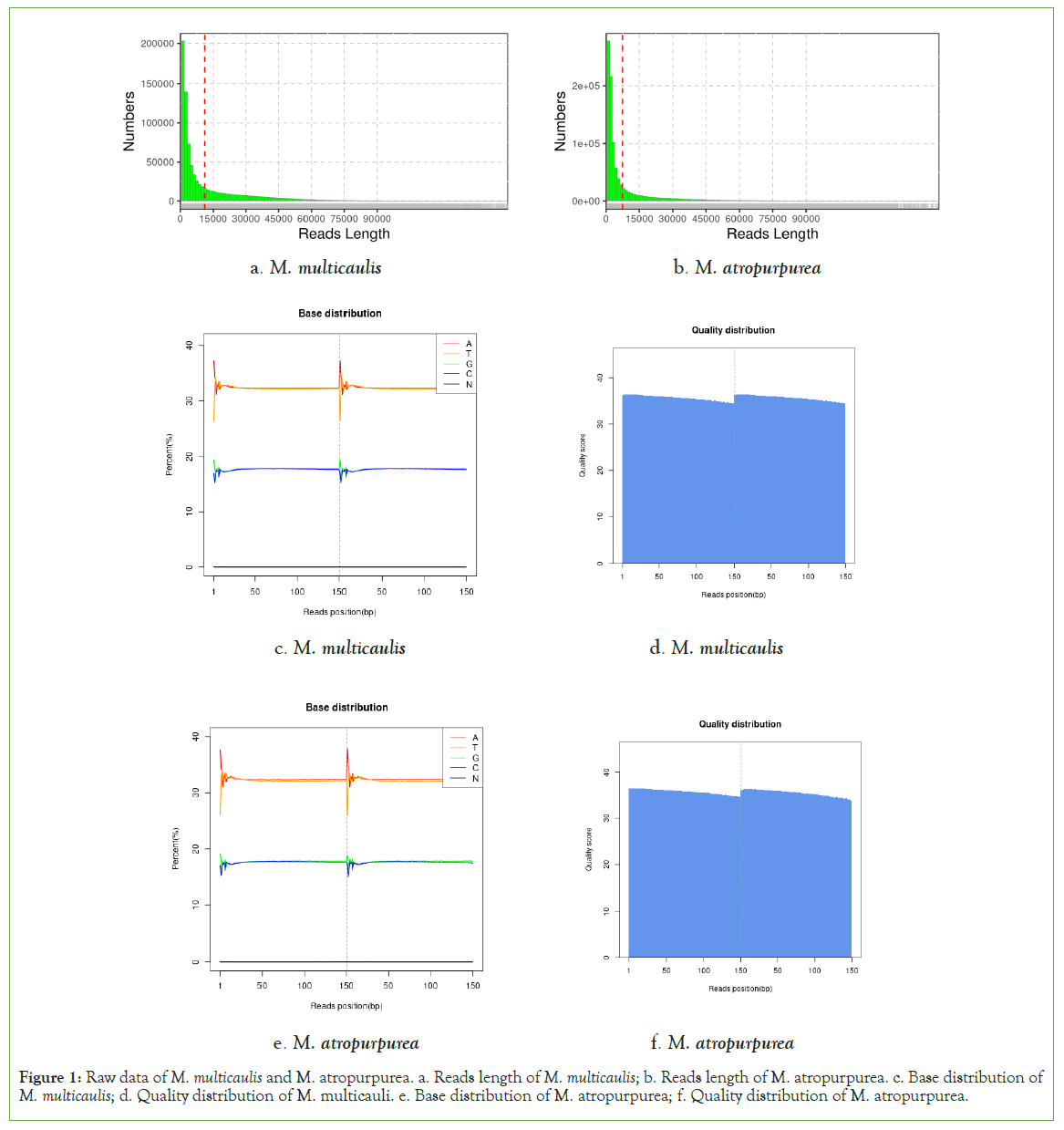
Figure 1: Raw data of M. multicaulis and M. atropurpurea. a. Reads length of M. multicaulis; b. Reads length of M. atropurpurea. c. Base distribution of M. multicaulis; d. Quality distribution of M. multicauli. e. Base distribution of M. atropurpurea; f. Quality distribution of M. atropurpurea.
| Material | Read sum | Base sum | GC (%) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|---|
| M. atropurpurea | 33780517 | 1.01E+10 | 35.36 | 96.88 | 91.87 |
| M. multicaulis | 29282471 | 8.78E+09 | 35.35 | 97.03 | 92.16 |
Table 1: Second generation sequencing data.
| Material | Number of reads | Number of bases | Mean read length | N50 read length |
|---|---|---|---|---|
| M. atropurpurea | 1112754 | 8140970643 | 7316 | 24118 |
| M. multicaulis | 930791 | 10385404567 | 11157 | 31464 |
Table 2: Third generation sequencing data.
Genome content and organization
The M. multicaulis mt genome is circular and was determined to be 361,546 bp long. The base composition of the genome is A (27.38%), T (27.20%), C (22.63%), G (22.79%), containing 54 functional genes. These include 3 rRNA genes, 20 tRNA genes, and 31 PCGs Pseudogenes and ORFs, which were all non-coding. The mt genome of M. multicaulis functional categorization and physical locations of the annotated genes were presented, encoding 31 different proteins that could be divided into 9 classes (Table 3) (Figure 2). Amongst these are ATP synthase (5 genes), cytochrome C biogenesis (4 genes), ubiquinol cytochrome C reductase (1 gene), Cytochrome C oxidase (3 genes), maturases (2 genes), transport membrane protein (1 gene), NADH dehydrogenase (9 genes), ribosomal proteins (SSU) (5 genes) and ribosomal proteins (LSU) (1 gene).
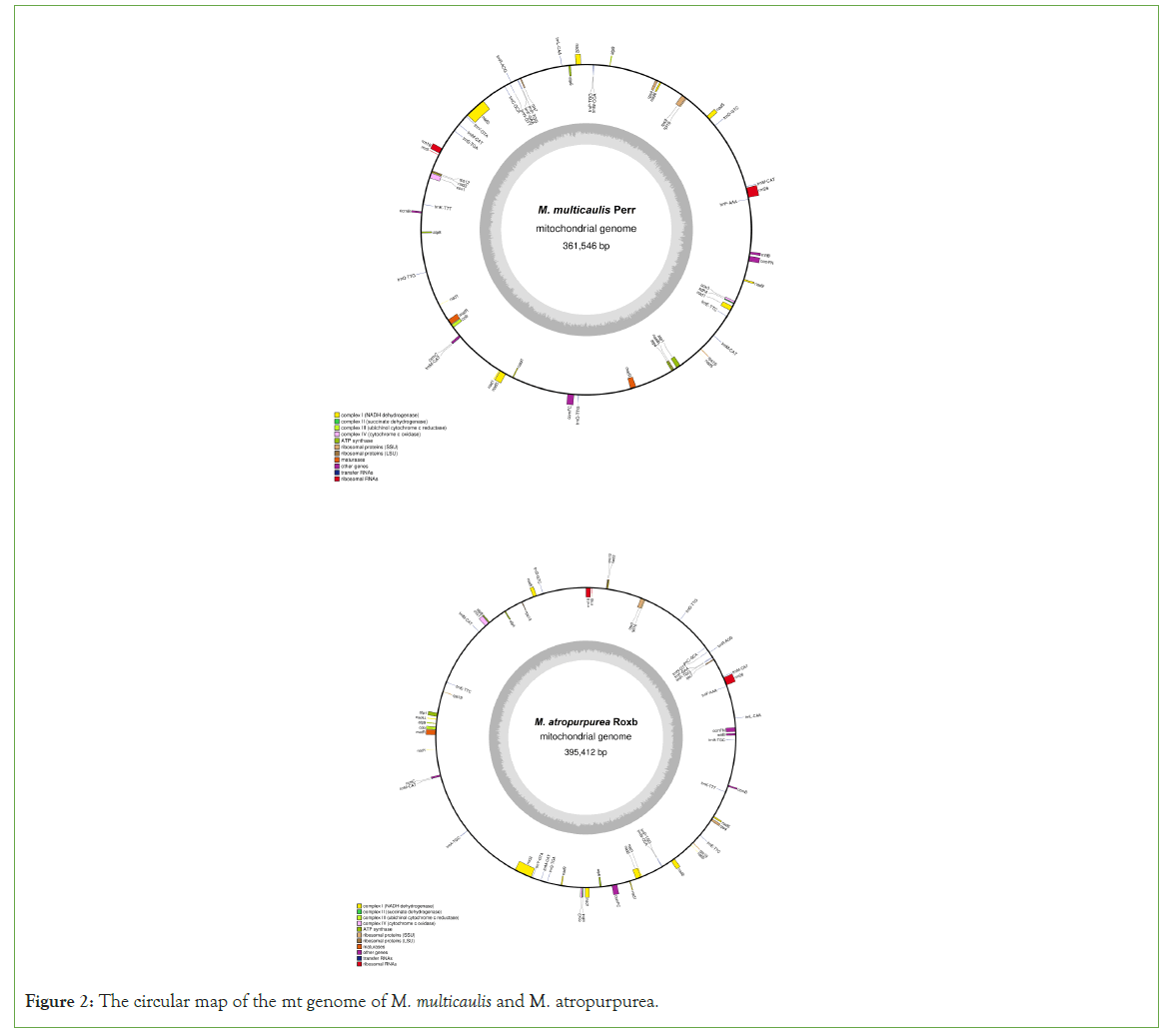
Figure 2: The circular map of the mt genome of M. multicaulis and M. atropurpurea.
| Characteristics | M. notabilis | M. multicaulis | M. atropurpurea |
|---|---|---|---|
| Size (bp) | 362,069 bp | 361546 | 395412 |
| GC content (%) | 45.66 | 45.42 | 45.50 |
| Number of genes | 54 | 54 | 57 |
| Protein-coding genes | 26 | 31 | 32 |
| rRNA | 3 | 3 | 3 |
| tRNA | 21 | 20 | 22 |
Table 3: Comparison of mt genomes among four species of Morus L.
The mt genome sequence of M. atropurpurea is also circular and found to be 395,412-bp long. The base comprises C+G (45.50%), including 57 functional genes containing 2 rRNA genes, 22 tRNA genes and 32 PCGs, 31 different proteins, divided into 9 classes (Table 4).
| Group of genes | M. multicaulis | M. atropurpurea |
|---|---|---|
| ATP synthase | atp1 atp4 atp6 atp8 atp9 | atp1 atp4 atp6 atp8 atp9 |
| Cytohrome c biogenesis | ccmB ccmC ccmFC* ccmFN | ccmB ccmC ccmFC* ccmFN |
| Ubichinol cytochrome creductase | cob | Cob |
| Cytochrome c oxidase | cox1 cox2* cox3 | cox1 cox2* cox3 |
| Maturases | matR(2) | matR |
| Transport membrance protein | mttB | mttB |
| NADH dehydrogenase | nad1**** nad2**** nad3 nad4** nad4L nad5**** nad6 nad7**** nad9 | nad1**** nad2**** nad3 nad4** nad4L nad5**** nad6 nad7**** nad9 |
| Ribosomal proteins (LSU) | rpl16 | rpl16 |
| Ribosomal proteins (SSU) | rps12 rps19 rps3 rps4 rps7 | rps12 rps13 rps19(2) rps3 rps4 rps7 |
| Succinate dehydrogenase | ψsdh4 | ψsdh4 |
| Ribosomal RNAs | rrn18 rrn26 rrn5 | rrn18 rrn26 rrn5 |
| Transfer RNAs | trnC-GCA trnD-GTC trnE-TTC trnF-AAA* trnF-GAA trnK-TTT trnL-CAA trnM-CAT(4) trnN-GTT trnP-TGG(2) trnQ-TTG(2) trnR-ACG trnS-TGA trnW-CCA trnY-GTA | trnA-TGC*(2) trnC-GCA trnD-GTC trnE-TTC(2) trnF-AAA* trnF-GAA trnK-TTT trnL-CAA trnM-CAT(4) trnN-GTT trnP-TGG(2) trnQ-TTG trnR-ACG trnS-TGA trnW-CCA trnY-GTA |
Note: The numbers after the gene names indicate the duplication number. “*” indicate genes containing one or more introns. “ψ” indicate pseudogene.
Table 4: Genes present in the mt genome of M. atropurpurea and M. multicaulis.
Variations and codon usage
In this study, the mt genome of M multicaulis and M. atropurpurea 27,933 and 28,251 codons, respectively. For M. multicaulis, 31 protein-coding genes in the mt genome were encoded by 27,933 codons. The codon end at A or T accounted for 62.2%. Leu accounts for the highest codon usage (3,084), followed by Ser (2,454) and Arg (1,824) (Figures 3). These three amino acids almost represent four-fifths of the total codons. The codon with the least number is Trp (459). All the protein-coding genes used AUG (753) as the most common start codon and three stop codons UAA, UGA, and UAG with the following utilization rate: UAA (53.33%), UGA (23.33%), and UAG (23.33%). The mitochondrial genomes of M. atropurpurea were encoded by 28,251 codons. Among them, the most coding codon was leucine (Leu) 3,096, followed by serine (2,481) and Arginine (1,842), and the least number is Trp (456).
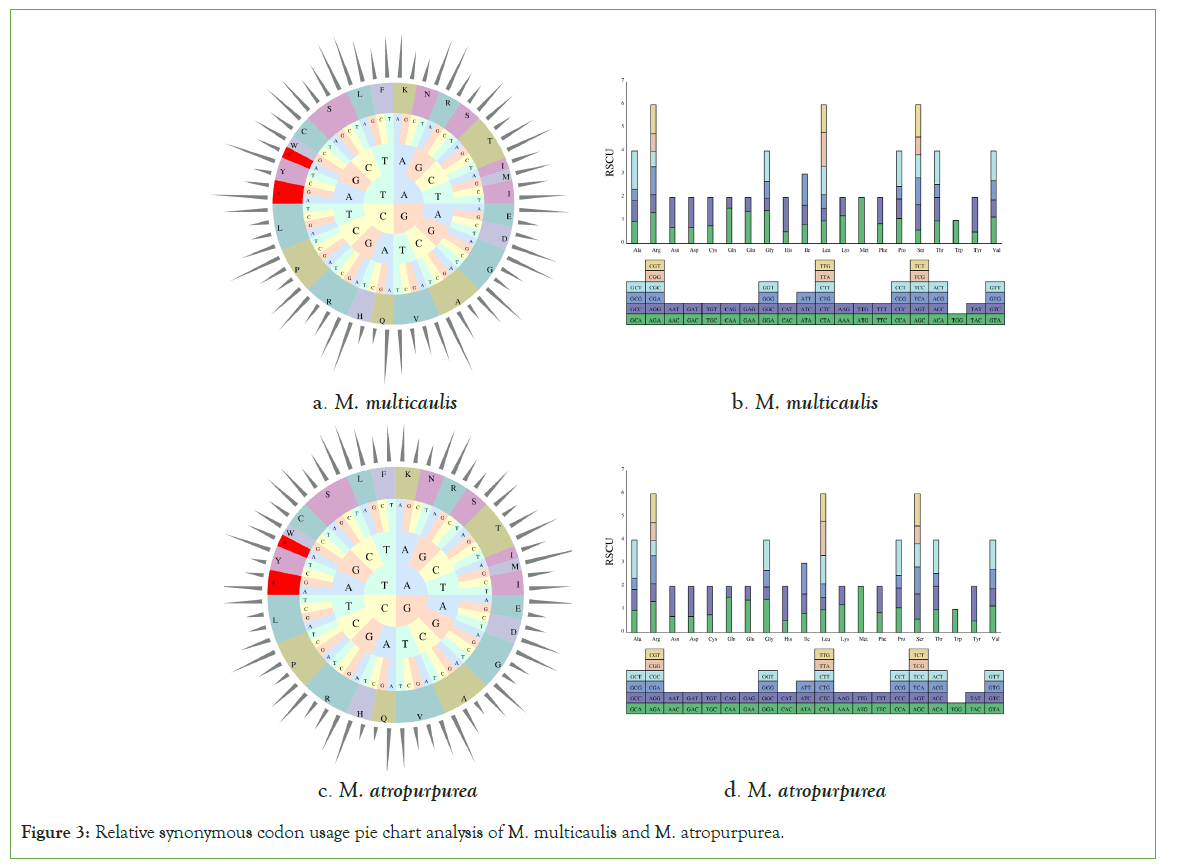
Figure 3: Relative synonymous codon usage pie chart analysis of M. multicaulis and M. atropurpurea.
Previous reports have shown that the mt genomes contain a variable number of introns [15]. In our results the mt genome of M. multicaulis has 8 intron-containing genes (ccmFC, cox2, nad1, nad2, nad4, nad5, nad7, trnF-AAA) harboring 21 introns in total. Moreover, nad1, nad2, nad5, nad7 contains 4 introns, which is the highest intron number. On the hand, M. atropurpurea had 8 intron-containing genes comprising 21 introns. Most land plants contain 3 rRNA genes [16]. Consistently, in our study, two species contain 3 rRNA genes (rrn18, rrn26 and rrn5) thus were annotated in Morus mt genome. Moreover, 20 different RNAs transfer were identified in the M. multicaulis mt genome transporting 19 amino acids, which indicate that more than one RNAs transfer might occur in the same amino acid with different codons.
Repeat sequences analysis
Tandem repeats, also named satellite DNA, are widely found in eukaryotic and some prokaryotes genomes (GAO H 2005). Scattered repetitive sequences are another type of repetitive sequence different from tandem repetitive sequences, distributed in a dispersed manner in the genome. We use vmatch v2.3.0 software to identify as follows: forward, palindromic, reverse, and complement. It was shown that the 30-40 bp repeats were most abundant in both species. In the mitochondrial genome of M. multicaulis, there were 53 scattered repeats, accounting for 8.93% of the total length, and the longest repeat is 22,003 bp. Also, M. atropurpurea had 69 scattered repeats, accounting for 2.04% of the total length with the longest repeat being 20,931 bp (Figure 4).
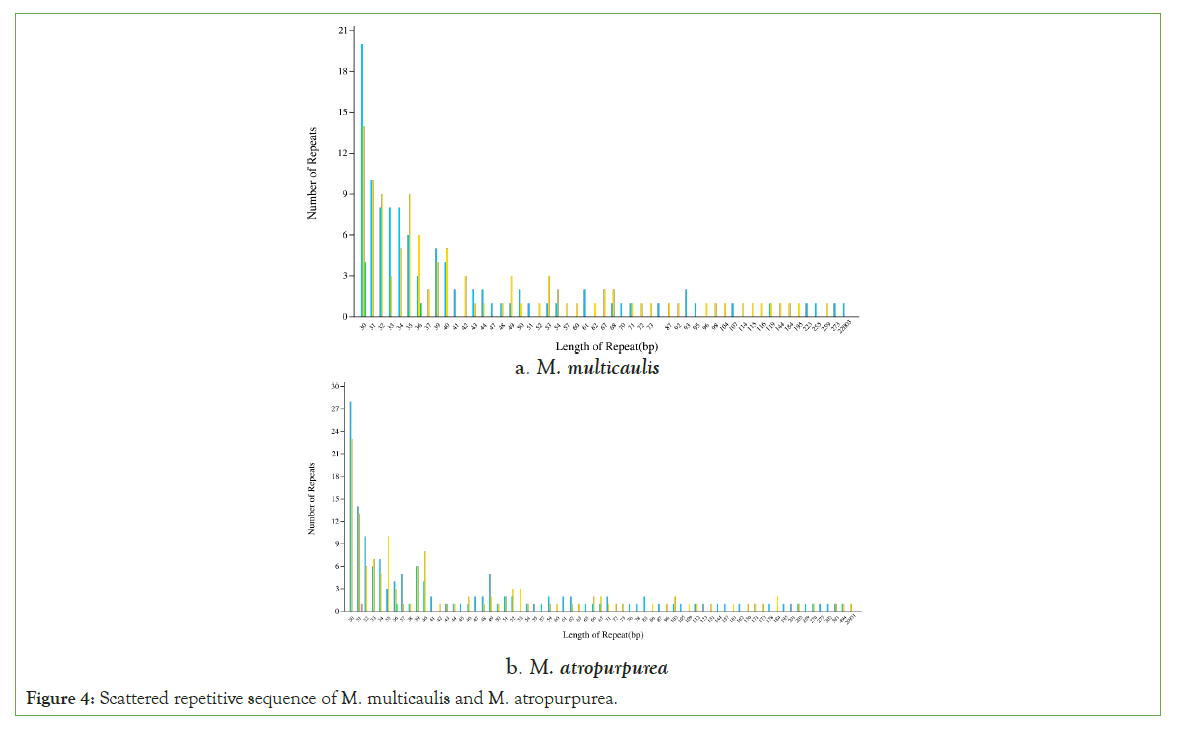
Figure 4: Scattered repetitive sequence of M. multicaulis and M. atropurpurea.
The prediction of RNA editing
RNA editing refers to the addition, loss, and conversion of the exist in the transcribed RNA's coding region found in all eukaryotes and plants [17,18]. The conversion of specific cytosine into uridine can alter genomic information, has been reported [19]. In this study, we used online sites (http://www.prepact.de/prepact-main.php) to predict the RNA editing sites. The results showed that a total of 377 RNA editing sites within 22 protein-coding genes were identified in M. multicaulis. Interestingly, mttB, nad5 and ccmC were the most editing sites predicted (32). There were 8 protein-coding genes (atp1, atp6, atp8, cox1, cox2, cox3, rpl16, rps19) which do not have any editing site predicted in the mt genome of M. multicaulis. According to the results, among those editing sites, 36.07% (136) occurred at the first base of the triplet position and 63.93% (241) were located at the second base of the triplet position. The hydrophobicity percentage indicates that 42.18% of amino acids did not change. However, 9.28% of the amino acids were predicted to change from hydrophobic to hydrophilic, and 48.54% changed from hydrophilic to hydrophobic (Table 5).
| Type | Codon | Aa change | Number | Percentage |
|---|---|---|---|---|
| Hydrophobic | TTT->CTT | F->L | 4 | 28.12% |
| TTG->CTG | F->L | 3 | ||
| GCT->GTT | A->V | 1 | ||
| GCG->GTG | A->V | 2 | ||
| GCA->GTA | A->V | 1 | ||
| CTT->TTT | L->F | 11 | ||
| CTC->TTC | L->F | 3 | ||
| CCT->CTT | P->L | 20 | ||
| CCG->CTG | P->L | 19 | ||
| CCC->CTC | P->L | 8 | ||
| CTC->CCC | L->P | 1 | ||
| CCA->CTA | P->L | 33 | ||
| Hydrophilic | CGT->TGT | R->C | 23 | |
| CGC->TGC | R->C | 9 | 14.06% | |
| CAT->TAT | H->Y | 13 | ||
| CAC->TAC | H->Y | 8 | ||
| Hydrophobic-hydrophilic | CCT->TCT | P->S | 17 | |
| CCG->TCG | P->S | 5 | 9.28% | |
| CCC->TCC | P->S | 11 | ||
| CCA->TCA | P->S | 2 | ||
| Hydrophilic-hydrophobic | TCT->TTT | S->F | 34 | |
| TCG->TTG | S->L | 62 | ||
| TCA->TTA | S->L | 49 | 48.54% | |
| ACT->ATT | T->I | 4 | ||
| ACG->ATG | T->M | 3 | ||
| ACC->ATC | T->I | 1 | ||
| ACA->ATA | T->I | 3 | ||
| TCC->CCC | S->P | 1 | ||
| TCA->CCA | S->P | 2 | ||
| CGG->TGG | R->W | 24 |
Table 5: Prediction of RNA editing sites of M. multicaulis.mt genome.
In the mitochondrial genome of M. atropurpurea, 373 RNA editing sites were found in 23 protein-coding genes. Within this, 36.46% (136) located at the first position of the triplet position and 63.54% (237) occurred at the second base of triplet position. Here also, mttb, nad5 and ccmC were the most RNA editing sites (32) and the least with only one editing site being nad4l (Figure 5). Furthermore, 42.44% the M. atropurpurea amino acids had no change in hydrophobicity, 48.53% and 9.38% of the amino acids were changed from hydrophilicity to hydrophobicity, and hydrophobicity to hydrophilicity, respectively (Table 6).
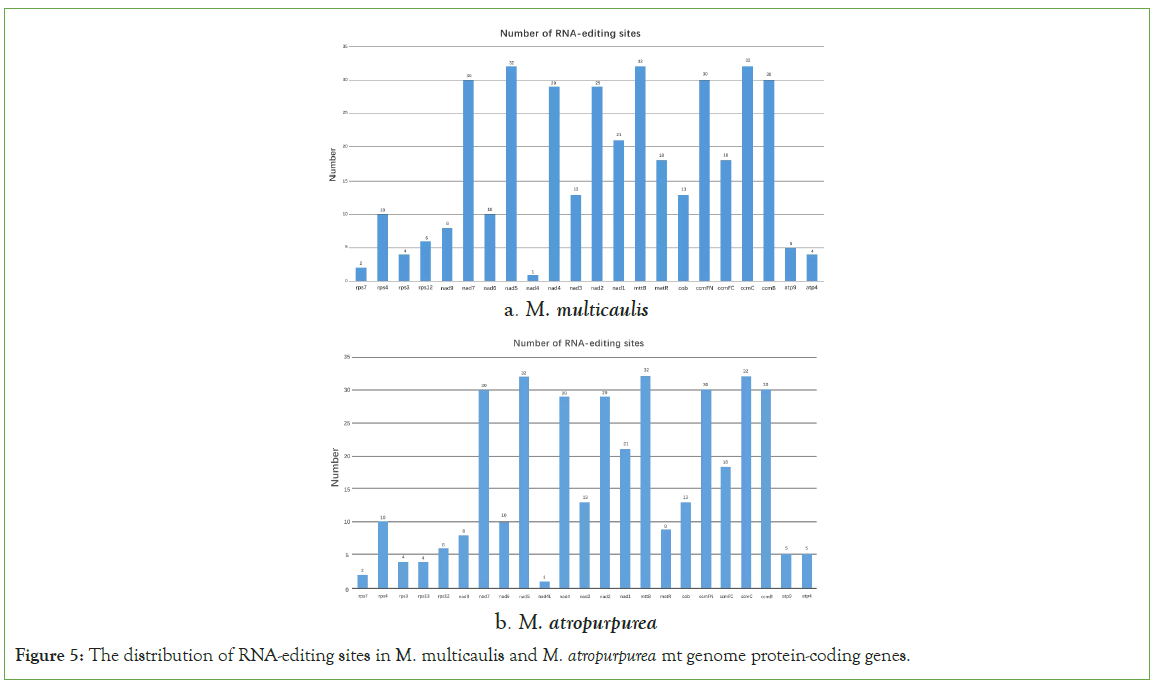
Figure 5: The distribution of RNA-editing sites in M. multicaulis and M. atropurpurea mt genome protein-coding genes.
| Type | Codon | Aa change | Number | Percentage |
|---|---|---|---|---|
| Hydrophobic | CCC->CTC | P->L | 7 | 28.15% |
| CCA->CTA | P->L | 32 | ||
| CCG->CTG | P->L | 19 | ||
| CTC->CCC | L->P | 1 | ||
| CTC->TTC | L->F | 3 | ||
| CTT->TTT | L->F | 11 | ||
| GCA->GTA | A->V | 1 | ||
| GCG->GTG | A->V | 2 | ||
| GCT->GTT | A->V | 1 | ||
| TTC->CTC | F->L | 3 | ||
| TTG->CTG | F->L | 1 | ||
| TTT->CTT | F->L | 5 | ||
| CCT->CTT | P->L | 19 | ||
| Hydrophilic | CAC->TAC | H->Y | 7 | 13.94% |
| CAT->TAT | H->Y | 13 | ||
| CGC->TGC | R->C | 8 | ||
| CGT->TGT | R->C | 24 | ||
| Hydrophobic-hydrophilic | CCA->TCA | P->S | 2 | 9.38% |
| CCC->TCC | P->S | 11 | ||
| CCT->TCT | P->S | 17 | ||
| CCG->TCG | P->S | 5 | ||
| Hydrophilic-hydrophobic | ACA->ATA | T->I | 3 | 48.53% |
| ACC->ATC | T->I | 1 | ||
| ACG->ATG | T->M | 3 | ||
| ACT->ATT | T->I | 4 | ||
| CGG->TGG | R->W | 24 | ||
| TCA->CCA | S->P | 1 | ||
| TCA->TTA | S->L | 49 | ||
| TCC->CCC | S->P | 1 | ||
| TCC->TTC | S->F | 27 | ||
| TCG->TTG | S->L | 34 | ||
| TCT->TTT | S->F | 34 |
Table 6: Prediction of RNA editing sites of M. atropurpurea mt genome.
Homology analysis of chloroplast with mitochondria
DNA migration is common in plants [20]. The homologous sequence between chloroplast and mitochondria was found using blast software. The similarity was set to 70% and e-value to 10E-5 using circos v0.69-5 to visualize it. Twenty-five fragments with a total length of 28,207 bp were observed to be migrated from the cp genome to the mt genome in M. multicaulis, accounting for 7.80% of the mt genome (Figures 6). Seven annotated genes were identified on those fragments, tRNA genes: namely trnL-CAA, trnN-GTT, trnM-CAT, trnP-TGG, trnW-CCA, trnD-GTC, and trnM-CAT (Table 7). In the M. atropurpurea, 44 fragments with a total length of 33834 bp were observed, accounting for 8.56% of the total length. About seven tRNA genes: trnL-CAA,trnN-GTT,trnA-TGC,trnM-CAT,trnP-TGG,trnW-CCA,trnD-GTC were identified (Table 8).
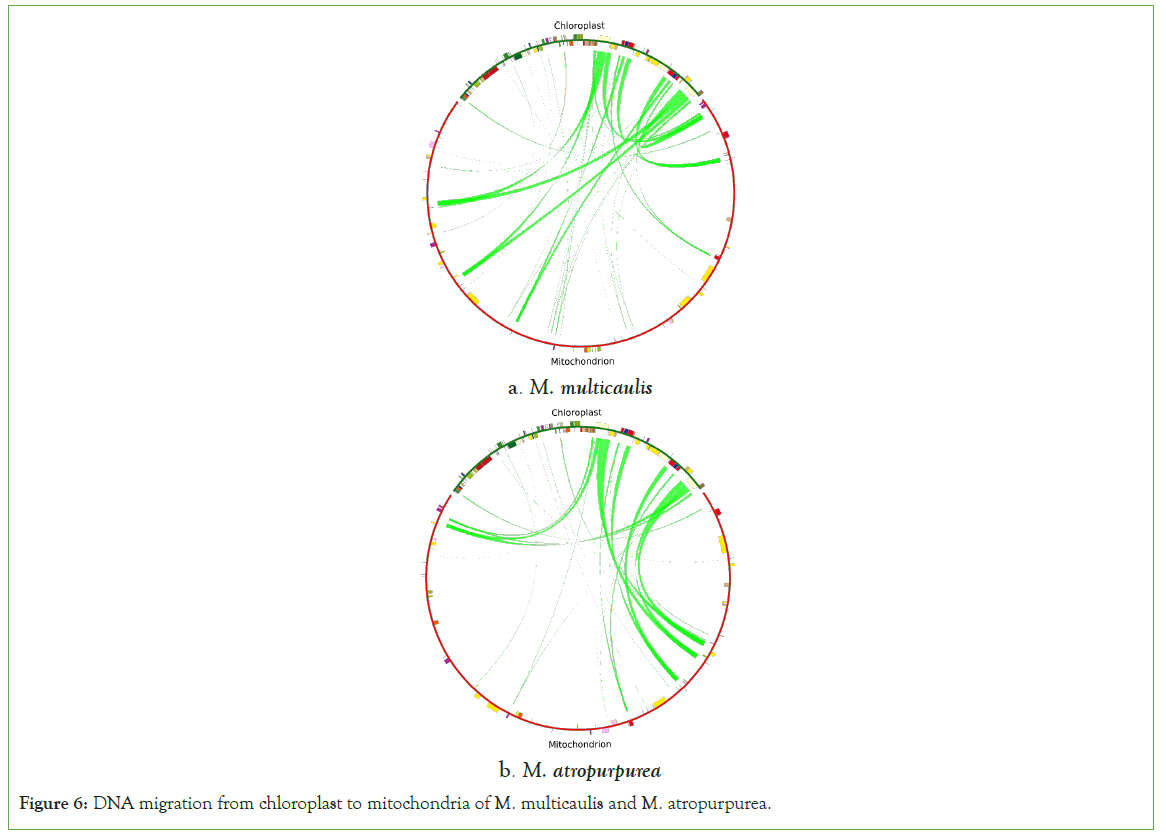
Figure 6: DNA migration from chloroplast to mitochondria of M. multicaulis and M. atropurpurea.
| S.no | length | identity | Mis match | Gap opens | mt start | mt end | cp start | cp end | Gene |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 3112 | 98.747 | 13 | 3 | 91,640 | 88,555 | 150,948 | 154,059 | |
| 2 | 3112 | 98.747 | 13 | 3 | 88,555 | 91,640 | 93,119 | 96,230 | |
| 3 | 2936 | 99.251 | 8 | 1 | 100,235 | 97,314 | 96,350 | 99,285 | |
| 4 | 2936 | 99.251 | 8 | 1 | 97,314 | 100,235 | 147,893 | 150,828 | trnL-CAA |
| 5 | 2681 | 99.925 | 1 | 1 | 118,160 | 115,481 | 134,823 | 137,503 | trnL-CAA |
| 6 | 2681 | 99.925 | 1 | 1 | 115,481 | 118,160 | 109,675 | 112,355 | trnN-GTT |
| 7 | 2180 | 99.083 | 11 | 1 | 346,348 | 344,178 | 91,029 | 93,208 | trnN-GTT |
| 8 | 2180 | 99.083 | 11 | 1 | 344,178 | 346,348 | 153,970 | 156,149 | |
| 9 | 1073 | 98.788 | 5 | 1 | 349,695 | 348,631 | 88,408 | 89,480 | |
| 10 | 1073 | 98.788 | 5 | 1 | 348,631 | 349,695 | 157,698 | 158,770 | |
| 11 | 521 | 87.716 | 18 | 9 | 7,055 | 7,529 | 796 | 1,316 | |
| 12 | 235 | 100 | 0 | 0 | 221,648 | 221,414 | 89,864 | 90,098 | |
| 13 | 235 | 100 | 0 | 0 | 221,414 | 221,648 | 157,080 | 157,314 | nad1*,ccmC,trnM-CAT |
| 14 | 889 | 74.241 | 174 | 42 | 152,321 | 151,463 | 141,980 | 142,843 | nad1*,ccmC,trnM-CAT |
| 15 | 889 | 74.241 | 174 | 42 | 151,463 | 152,321 | 104,335 | 105,198 | rrn18 |
| 16 | 507 | 79.29 | 67 | 25 | 87,604 | 88,091 | 69,809 | 70,296 | rrn18 |
| 17 | 166 | 100 | 0 | 0 | 84,192 | 84,027 | 104,835 | 105,000 | trnP-TGG,trnW-CCA |
| 18 | 166 | 100 | 0 | 0 | 84,027 | 84,192 | 142,178 | 142,343 | |
| 19 | 156 | 92.308 | 7 | 2 | 122,916 | 123,067 | 59,360 | 59,514 | |
| 20 | 148 | 92.568 | 10 | 1 | 245,879 | 245,732 | 36,734 | 36,880 | |
| 21 | 82 | 97.561 | 1 | 1 | 39,222 | 39,142 | 32,340 | 32,421 | |
| 22 | 79 | 94.937 | 4 | 0 | 141,020 | 140,942 | 55,104 | 55,182 | trnD-GTC |
| 23 | 62 | 90.323 | 4 | 2 | 164,557 | 164,498 | 146,669 | 146,730 | trnM-CAT |
| 24 | 62 | 90.323 | 4 | 2 | 164,498 | 164,557 | 100,448 | 100,509 | |
| 25 | 46 | 95.652 | 0 | 1 | 326,639 | 326,596 | 45,875 | 45,920 | |
| Total | 28,207 |
Table 7: Fragments transferred from chloroplast to mitochondria of M. multicaulis mt genome.
| S.no | length | identity | Mis match | Gap opens | mt start | mt end | cp start | cp end | Gene |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 3112 | 99.197 | 8 | 2 | 333,927 | 330,833 | 150,963 | 154,074 | |
| 2 | 3112 | 99.197 | 8 | 2 | 330,833 | 333,927 | 93,163 | 96,274 | |
| 3 | 2936 | 99.046 | 9 | 2 | 9,427 | 6,511 | 96,394 | 99,329 | trnL-CAA |
| 4 | 2936 | 99.046 | 9 | 2 | 6,511 | 9,427 | 147,908 | 150,843 | trnL-CAA |
| 5 | 2681 | 99.925 | 1 | 1 | 38,649 | 35,970 | 134,838 | 137,518 | trnN-GTT |
| 6 | 2681 | 99.925 | 1 | 1 | 35,970 | 38,649 | 109,719 | 112,399 | trnN-GTT |
| 7 | 2509 | 99.243 | 10 | 1 | 285,883 | 283,384 | 153,985 | 156,493 | |
| 8 | 2509 | 99.243 | 10 | 1 | 283,384 | 285,883 | 90,744 | 93,252 | |
| 9 | 1668 | 100 | 0 | 0 | 240,840 | 239,173 | 138,989 | 140,656 | trnA-TGC |
| 10 | 1668 | 100 | 0 | 0 | 239,173 | 240,840 | 106,581 | 108,248 | trnA-TGC |
| 11 | 949 | 100 | 0 | 0 | 6,058 | 5,110 | 157,837 | 158,785 | |
| 12 | 949 | 100 | 0 | 0 | 5,110 | 6,058 | 88,452 | 89,400 | |
| 13 | 521 | 87.716 | 18 | 9 | 18,239 | 18,713 | 796 | 1,316 | |
| 14 | 216 | 100 | 0 | 0 | 214,334 | 214,119 | 89,927 | 90,142 | ccmC/trnM-CAT |
| 15 | 216 | 100 | 0 | 0 | 214,119 | 214,334 | 157,095 | 157,310 | ccmC/trnM-CAT |
| 16 | 230 | 96.087 | 0 | 1 | 217,316 | 217,096 | 156,145 | 156,374 | |
| 17 | 230 | 96.087 | 0 | 1 | 217,096 | 217,316 | 90,863 | 91,092 | |
| 18 | 889 | 74.241 | 174 | 42 | 98,535 | 97,677 | 104,379 | 105,242 | rrn18 |
| 19 | 889 | 74.241 | 174 | 42 | 97,677 | 98,535 | 141,995 | 142,858 | rrn18 |
| 20 | 506 | 79.249 | 69 | 23 | 329,882 | 330,369 | 69,854 | 70,341 | trnP-TGG/trnW-CCA |
| 21 | 166 | 100 | 0 | 0 | 169,697 | 169,532 | 142,193 | 142,358 | |
| 22 | 166 | 100 | 0 | 0 | 169,532 | 169,697 | 104,879 | 105,044 | |
| 23 | 166 | 100 | 0 | 0 | 355,190 | 355,025 | 104,879 | 105,044 | |
| 24 | 166 | 100 | 0 | 0 | 355,025 | 355,190 | 142,193 | 142,358 | |
| 25 | 143 | 98.601 | 1 | 1 | 165,372 | 165,231 | 157,434 | 157,576 | |
| 26 | 143 | 98.601 | 1 | 1 | 165,231 | 165,372 | 89,661 | 89,803 | |
| 27 | 156 | 92.308 | 7 | 2 | 211,340 | 211,189 | 59,407 | 59,561 | |
| 28 | 148 | 92.568 | 10 | 1 | 314,042 | 314,189 | 36,770 | 36,916 | |
| 29 | 112 | 100 | 0 | 0 | 219,737 | 219,626 | 89,413 | 89,524 | |
| 30 | 112 | 100 | 0 | 0 | 219,626 | 219,737 | 157,713 | 157,824 | |
| 31 | 108 | 98.148 | 2 | 0 | 214,437 | 214,330 | 157,550 | 157,657 | |
| 32 | 108 | 98.148 | 2 | 0 | 214,330 | 214,437 | 89,580 | 89,687 | |
| 33 | 108 | 98.148 | 2 | 0 | 245,900 | 245,793 | 157,550 | 157,657 | |
| 34 | 108 | 98.148 | 2 | 0 | 245,793 | 245,900 | 89,580 | 89,687 | |
| 35 | 82 | 97.561 | 1 | 1 | 117,087 | 117,007 | 32,352 | 32,433 | trnD-GTC |
| 36 | 79 | 94.937 | 4 | 0 | 277,436 | 277,358 | 55,161 | 55,239 | trnM-CAT |
| 37 | 62 | 90.323 | 4 | 2 | 364,620 | 364,561 | 146,684 | 146,745 | |
| 38 | 62 | 90.323 | 4 | 2 | 364,561 | 364,620 | 100,492 | 100,553 | |
| 39 | 46 | 95.652 | 0 | 1 | 177,030 | 177,073 | 45,933 | 45,978 | |
| 40 | 46 | 95.652 | 0 | 1 | 347,692 | 347,649 | 45,933 | 45,978 | |
| 41 | 37 | 100 | 0 | 0 | 245,773 | 245,737 | 157,293 | 157,329 | |
| 42 | 37 | 100 | 0 | 0 | 245,737 | 245,773 | 89,908 | 89,944 | |
| 43 | 33 | 96.97 | 1 | 0 | 245,797 | 245,765 | 89,927 | 89,959 | |
| 44 | 33 | 96.97 | 1 | 0 | 245,765 | 245,797 | 157,278 | 157,310 | |
| Total | 33,834 |
Table 8: Fragments transferred from chloroplast to mitochondria of M. atropurpurea mt genome
Our data demonstrate that some chloroplast protein-coding genes migrated from cp to mitochondrion. Most of them lost their integrities during evolution, and only partial sequences of those genes could be found in the mt genome, such as nad1 ccmC, rrn18. The different destinations of transferred protein-coding genes and tRNA genes suggested that the tRNA gene is much more conserved in the mt genome than the protein-coding genes, indicating their indispensable roles in mitochondria.
Comparison with other green plant mt genomes
The mulberry mt genome sequence was compared with other plastomes at the global level using mVISTA online software in the shuffle-LAGAN mode. Morus species with four families (Leguminosae, Gramineae, Rosaceae, Asteraceae) were used. M. notabilis was used as the reference in the comparative analysis. Interestingly, four families were remarkably group-specific. Each group shows nearly identical patterns among themselves. M. multicaulis and M. atropurpurea were remarkably close, and both were clustered with Morus notabilis which means that they have a very close genetic relationship to Morus notabilis (Figure 7).
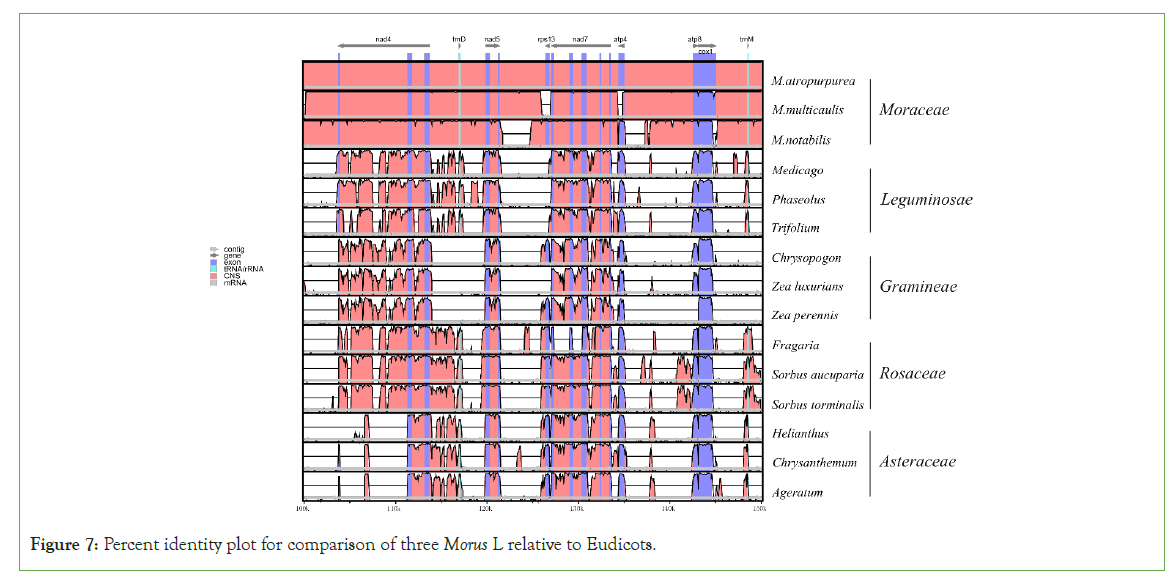
Figure 7: Percent identity plot for comparison of three Morus L relative to Eudicots.
Variation architecture at the mt genome level
Nucleic acid diversity (pi) can reveal the variation of nucleic acid sequences of different species and regions with higher variability. Thus, it provides potential molecular markers for population genetics. We use maft software (set at the auto mode) to compare the homologous gene sequences of distinct species globally and dnasp5 to calculate each gene's pi value. In the mt genome the nucleotide diversity (pi) of the mt genome in cultivated species, M. multicaulis with wild M. notabilis was calculated. We found 10 gene (cox1, ccmF, cob, ccmFN, nad9, mttB, nad3, nad4, atp4, atp9, and rps3) pi ranged from 0.00063 to 0.02182 slide window among whole mt genome. Most of the pi values were lower than 0.01, while rps3 accounting for the highest with 0.02182. In the M. atropurpurea, we identified 13 genes (ccmFc,cob,ccmFN,nad9,mttB,nad3,rps13,cox1,nad4,atp9,atp4,rps3,rps19). The gene rps19 was found to have the highest pi value (0.06343) (Figures 8). Furthermore, 85 variations, including 79 SNPs and 6 indels, were identified across the mt genomes of M. multicaulis and M. atropurpurea (Table 9). This phenomenon could be applied to analyze Morus mt genomic evolution further.
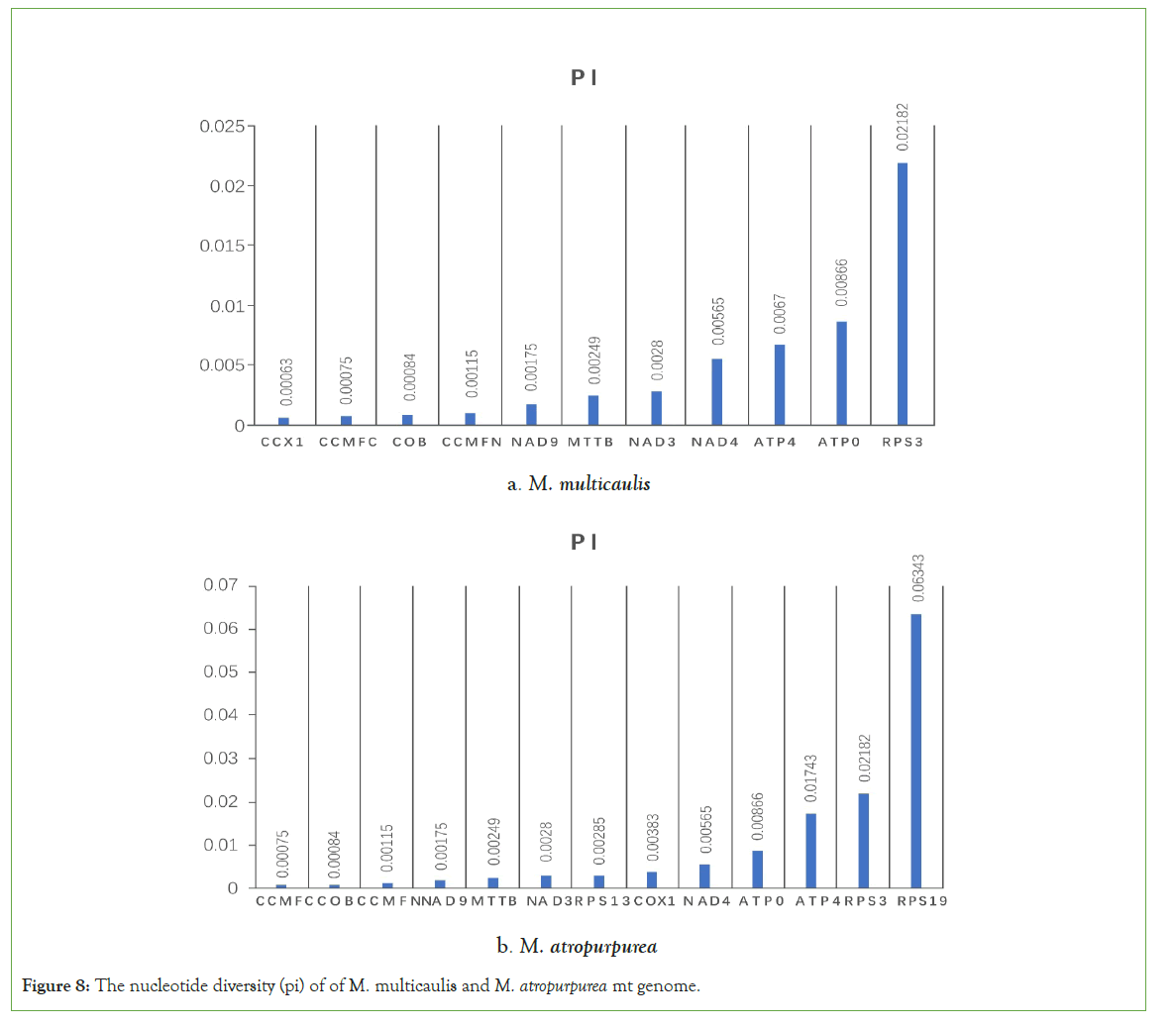
Figure 8: The nucleotide diversity (pi) of of M. multicaulis and M. atropurpurea mt genome.
| Summary | Type | Total variation |
|---|---|---|
| SNPs | 79 | |
| Indels | 6 | |
| Total | 85 |
Table 9: Summary the total variations (SNPs and Indels) in M. multicaulis and M. atropurpurea.
Phylogenetic analysis within dicotyledon mt genomes
To understand the evolutionary status of Morus, we use MEGA (7.0) to analyze Moraceae together with other 7 dicotyledons. A total of 28 species based on the complete mt genome sequence was selected. A phylogenetic tree was constructed through the ML and NJ methods with a bootstrap of 1,000 replicates to assess the reliability. The 28 eudicots selected from 8 families (Moraceae, Leguminosae, Gramineae, Brassicaceae, Malvaceae, Cucurbitaceae, Asteraceae. Solanaceae) were well clustered. The results showed that the phylogenetic tree strongly supports the order of taxa in the phylogenetic tree. This was consistent with those species' evolutionary relationships, indicating traditional taxonomy consistency with the molecular classification. Based on the phylogenetic relationships among the 28 species, different groups of plants can be applied to do further comparative analysis. For the Moraceae, all the two methods (ML and NJ) showed that M. atropurpurea and M. multicaulis were grouped. Thus, it revealed that M. atropurpurea and M. multicaulis are more related to their congeners than others. This analysis is important for the mt genome project, the development of molecular markers for Morus species (Figures 9 and 10).
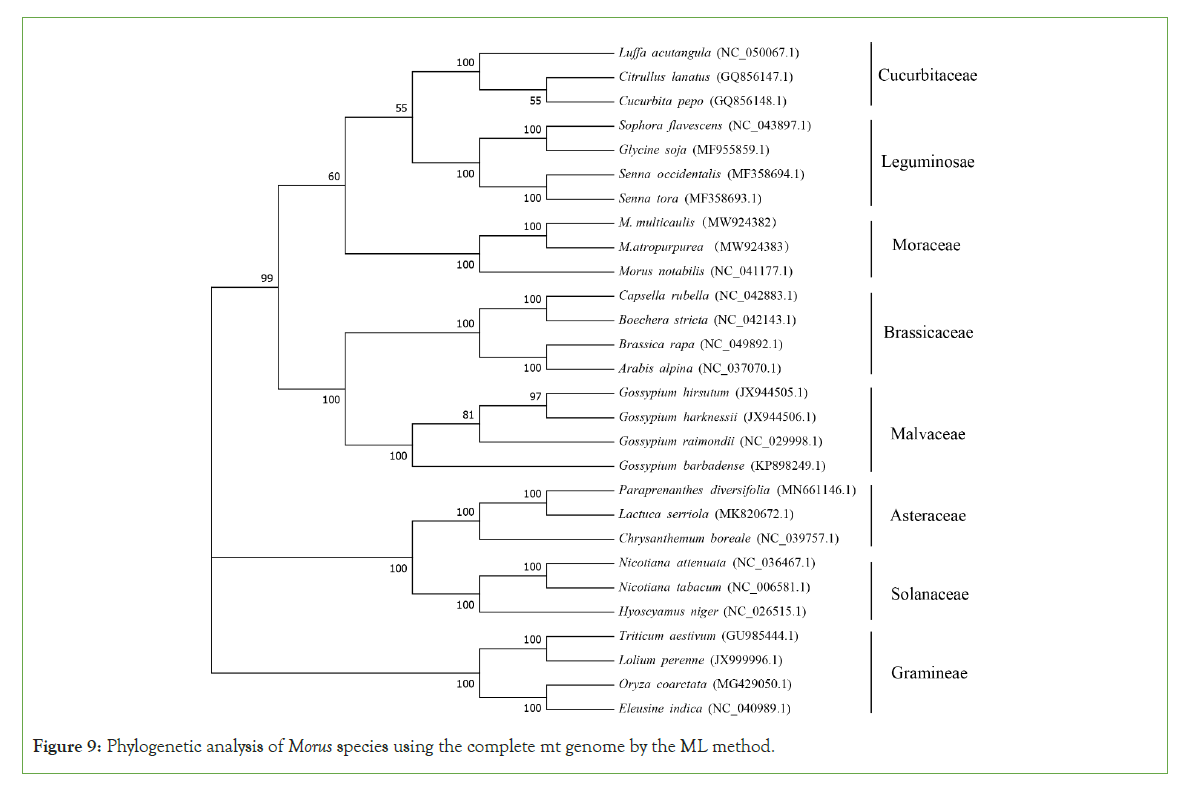
Figure 9: Phylogenetic analysis of Morus species using the complete mt genome by the ML method.
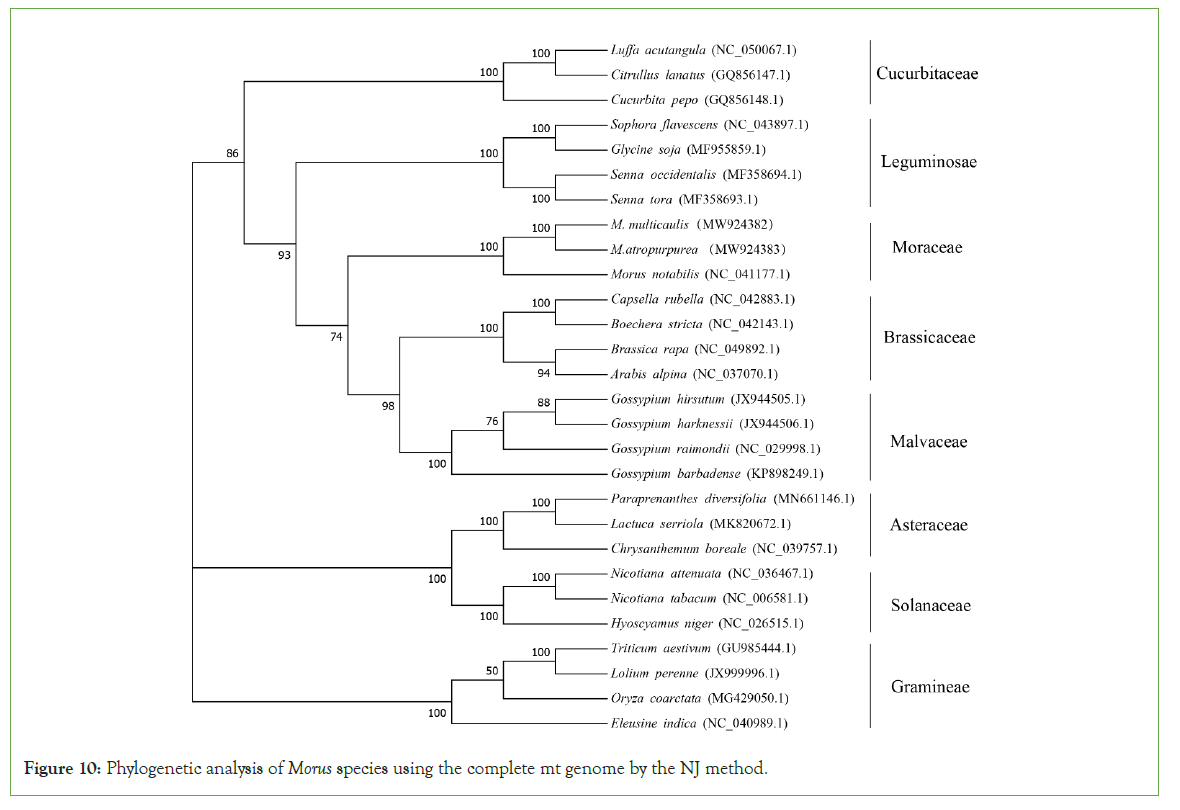
Figure 10: Phylogenetic analysis of Morus species using the complete mt genome by the NJ method.
Discussion
Mitochondria are the power source of energy required by plants to carry out life processes. It accounts for extensive size variations, sequence arrangements, repeat content, and highly conserved coding sequence, which are more complex than animals [2]. In this present study, we studied the mt genome's characteristics of mulberry. It is reported that most of the mt genome is circular, and few are linear such as the mt genome of Polytomella parva [21,22]. In the present study, the mt genome of M. multicaulis and M. atropurpurea is circular with 361,546 bp and 395,412 bp in size, respectively. The GC content of the mt genome Morus revels that GC content is highly conserved in higher plants.
The repeat sequences contain tandem, short, and large repeats that widely exist in the mt genome, thus, it plays a vital role in shaping the mt genome accounting for those repeats in mitochondria that are pivotal for intermolecular recombination [23]. We focus on reported scattered repetitive sequences intensively. Research has shown that M. multicaulis and M. atropurpurea harbors abundant repeat sequences that might indicate that the intermolecular recombination frequently happens in the mt genome, which may dynamically change the sequence and its conformation during evolution.
RNA-editing is a post-transcriptional process in both cpDNA and mt genomes of higher plants, contributing to the better folding of proteins [24]. Identifying RNA-editing sites provides essential clues for future analysis of predicting gene functions with novel codon about evolution. This can help better understand the gene expression of the cpDNA and mt genomes in plants. Previous studies have shown that Arabidopsis has a total of 441 RNA-editing sites within 36 genes, rice has 491 RNA-editing sites within 34 genes and 216 RNA-editing sites within 26 genes for S. glauca [14,21,25]. Our results show 377 RNA-editing sites within 22 protein-coding genes were predicted for M. multicaulis and 373 RNA-editing sites within 23 protein-coding genes were predicted for M. atropurpurea. The tRNA genes are much more conserved in the mt genome than the protein-coding genes, indicating their indispensable roles in mitochondria. As the cytoplasmic genome, migration of cpDNA to the mt genome occurred during the plant evolution. We found that 25 fragments were transferred from the cp genome to mt with 7 integrated genes, all tRNA genes. Transfer of tRNA genes from cp to mt is common in angiosperms [24]. Phylogenetic tree analysis indicates that M. atropurpurea and M. multicaulis are more related to their congeners than to others familiar [26]. Generally, most of the results in this study were consistent with previous reports.
Exploring and deciphering the mt genome is essential for plant breeding. Understanding the mt genome will set a foundation understanding for the evolutionary analysis, cytoplasmic male sterility, and molecular biological information evolution in mulberry plant [27-29].
In this study, we collected two cultivated species of Morus L. (M. atropurpurea and M. multicaulis), assembled, and annotated the mt genome and performed extensive analysis based on the complete mt genome sequences and amino acid sequences of the annotated genes [30-33]. We found out that the Morus species mt genome is circular, with M. multicaulis having a length of 361,546 bp. 54 genes, including 31 protein-coding genes, 20 tRNA genes, and 3 rRNA genes. Also, M. atropurpurea was found to have a length of 395,412bp. Moreover, a total of 57 genes contains 32 protein-coding genes, 22 tRNA and 3 rRNA were annotated in the genome.
Conclusion
The repeats sequences and RNA editing in M. multicaulis and M. atropurpurea mt genome were analyzed subsequently. The gene conversation between cpDNA and mt genome was also observed by detecting gene migration. Our result also indicates consistency in molecular and taxonomic classification, besides GC contents in angiosperms, were also found conserved despite their genome sizes that varied tremendously. This study provides extensive information about the mt genome for Morus L. It represents a valuable source of information for future studies on Morus populations and future breeding of Morus species
Statement
The experimental materials for this time are only experimental research and no field research. They were collected from my own school, National Mulberry GeneBank Zhenjiang. This collection was supported by Chinese Academy of Agricultural Sciences and with the guidance of the school official leaders. The collection was conducted under the conditions permitted by national laws and regulations; Strictly abide by relevant laws and get official permission. After the collection, it is only for experimental research and has no other purpose.
Acknowledgments
We thank National Mulberry GeneBank Zhenjiang for providing the plant sample of M. multicaulis and M.atropurpurea plant samples
Authors’ Contributions
Guo Liangliang and Zhao Weiguo conceived and designed the research. GL performed, assembled the genomes, analyzed the data, and wrote the original manuscript. Shi Yishu collected leaf samples, Wu Mengmeng extracted mitochondrial DNA. Michael Ackah revised the manuscript, LQ editing of the final manuscript; All authors contributed to the editing of the final manuscript.
Funding
This work was supported by China Agriculture Research System of MOF and MARA (CARS-18-ZJ0207), National Key R&D Program of China, key projects of international scientific and technological innovation cooperation (2021YFE0111100), Guangxi innovation-driven development project (AA19182012-2), Zhenjiang Science and Technology support project (GJ2021015).
Availability of Data and MaterialsThe sequence and annotation of M. multicaulis and M. atropurpurea mt genome data was deposited at the NCBI database with the MW924382 and MW924383 accession number in Gene Banks.
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare that they have no competing interests.
REFERENCES
- Chan WC, Lye PY, Wong SK. Phytochemistry, pharmacology and clinical trials of Morus alba. Chin J Nat Med. 2016;14(1):17-30.
- Kozik A, Rowan BA, Lavelle D, Berke L, Schranz ME, Christensen AC. The alternative reality of plant mitochondrial DNA: One ring does not rule them all. PLoS Genet. 2019;15:e1008373.
- Rehman J, Zhang HJ, Toth PT, Zhang Y, Marsboom G, Hong Z, et al. Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB J. 2012;26:2175-2186.
- Shtolz N, Dan. The mitochondrial genome-on selective constraints and signatures at the organism, cell, and single mitochondrion levels. Front Ecol Evol.2019;7:1.
- Yasunari O, Yukiko Y, Koji M, Akira K, Toru T, Takashi S, et al. Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Res. 2005;33(19):6235-6250.
- Richardson AO, Rice DW, Young GJ, Alverson AJ, Palmer JD. The "fossilized" mitochondrial genome of Liriodendron tulipifera: ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Bio. 2013;11:29.
- Tian X, Zheng J, Hu S, Yu J. The rice mitochondrial genomes and their variations. Plant Physiol. 2006;140(2):401-410.
- Sloan DB, Alverson AJ, Chuckalovcak JP, Wu M, McCauley DE, Palmer JD, et al. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012;10(1):e1001241.
- Skippington E, Barkman TJ, Rice DW, Palmer JD. Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc Natl Acad Sci USA. 2015;112(27):e3515-3524.
- Woloszynska M. Heteroplasmy and stoichiometric complexity of plant mitochondrial genomes--though this be madness, yet there's method in't. J Exp Bot. 2010;61(3):657-671.
- Alverson AJ, Xiaoxin W, Rice DW, Stern DB, Kerrie B, Palmer JD. Insights into the evolution of mitochondrial genome size from complete sequences of citrullus lanatus and cucurbita pepo (cucurbitaceae). Mol Biol Evol. 2010;27(6):1436-1448.
- Drouin G, Daoud H, Xia J. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol Phylogenet Evol. 2008;49(3):827-831.
- Tong W, Kim TS, Park YJ. Rice Chloroplast genome variation architecture and phylogenetic dissection in diverse oryza species assessed by whole-genome resequencing. Rice (N Y). 2016;9(1):57.
- Cheng Y, He X, Priyadarshani SVGN, Wang Y, Qin Y, Ye L, et al. Assembly and comparative analysis of the complete mitochondrial genome of Suaeda glauca. BMC Genomics. 2021;22(1):167.
- Liao X, Zhao Y, Kong X, Khan A, Bujin Z, Liu D, et al. Complete sequence of kenaf (Hibiscus cannabinus) mitochondrial genome and comparative analysis with the mitochondrial genomes of other plants. Sci Rep. 2018;8(1):12714.
- Archibald JM. Origin of eukaryotic cells: 40years on. Symbiosis. 2011;54:69-86.
- Kunzmann A, Brennicke A, Marchfelder A. 5' end maturation and RNA editing have to precede tRNA 3' processing in plant mitochondria. Proc Natl Acad Sci USA. 1998;95(1):108-113.
- Malek O, Lättig K, Hiesel R, Brennicke A, Knoop V. RNA editing in bryophytes and a molecular phylogeny of land plants. EMBO J. 1996;15(6):1403-1411.
- Rüdinger MS, Knoop V. Coevolution of organelle RNA editing and nuclear specificity factors in early land plants. ScienceDirect. 2016;78:37-93.
- Chang S, Wang Y, Lu J, Gai J, Li J, Chu P, et al. The mitochondrial genome of soybean reveals complex genome structures and gene evolution at intercellular and phylogenetic levels. PLoS One. 2013;8(2):e56502.
- Notsu Y, Masood S, Nishikawa T, Kubo N, Akiduki G, Nakazono M, et al. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol Genet Genomics. 2002;268(4):434-445.
- Smith DR, Lee RW. Mitochondrial genome of the colorless green alga polytomella capuana: a linear molecule with an unprecedented GC content. Mol Biol Evol. 2008;25(3):487-496.
- Dong S, Zhao C, Chen F, Liu Y, Zhang S, Wu H, et al. The complete mitochondrial genome of the early flowering plant Nymphaea colorata is highly repetitive with low recombination. BMC Genomics. 2018;19(1):614.
- Bi C, Paterson AH, Wang X, Xu Y, Wu D, Qu Y, et al. Analysis of the complete mitochondrial genome sequence of the diploid cotton Gossypium raimondii by comparative genomics approaches. Biomed Res Int. 2016;2016:5040598.
- Unseld M, Marienfeld JR, Brandt P, Brennicke A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat Genet. 1997;15(1):57-61.
- Li QL, Guo JZ, Yan N, Li CC. Complete chloroplast genome sequence of cultivated Morus L. species. Genet Mol Res. 2016;15(4).
- Gao H. Distribution characteristics and biological function of tandem repeat sequences in the genomes of different organisms. 2005;026:555-564. Google scholar]
- Guo W, Zhu A, Fan W, Mower JP. Complete mitochondrial genomes from the ferns Ophioglossum californicum and Psilotum nudum are highly repetitive with the largest organellar introns. New Phytol. 2017;213(1):391-403.
- He N, Zhang C, Qi X, Zhao S, Tao Y, Yang G, et al. Draft genome sequence of the mulberry tree Morus notabilis. Nat Commun. 2013;4:2445.
[Crossref][Google scholar][Pubmed ]
- Huang CY, Ayliffe MA, Timmis JN. Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature. 2003;422:72-76.
- Koren S, Walenz BP, Berlin K, Miller JR, Bergmann NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27(5):722-736.
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel M, Sakthikumar S, et al. Pilon: comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9(11):e112963.
- Zhang H, Li C, Miao H, Xiong S. Insights from the complete chloroplast genome into the evolution of Sesamum indicum L. PLoS One. 2013;8(11):e80508.
Citation: Liangliang G, Yisu S, Mengmeng W, Ackah M, Lin Q, Zhao W (2022) The Complete Mitochondrial Genome Sequence Variation and Phylogenetic Analysis of Mulberry”. J Data Mining Genomics Proteomics. 13:249.
Copyright: © 2022 Liangliang G, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

