Indexed In
- Academic Journals Database
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- ResearchBible
- Ulrich's Periodicals Directory
- Electronic Journals Library
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Commentary - (2020) Volume 11, Issue 2
Short Commentary on: On the Prediction of DNA-binding Preferences of C2H2-ZF Domains Using Structural Models: Application on Human CTCF
Alberto Meseguer1, Ruben Molina-Fernández1, Narcis Fernandez-Fuentes2, Oriol Fornes3* and Baldo Oliva1*2Department of Biosciences, U Science Tech, Universitat de Vic-Universitat Central de Catalunya, Vi, Catalonia 08500, Spain
3Centre for Molecular Medicine and Therapeutics, BC Children’s Hospital Research Institute, D, University of British Columbia, Vancouver, BC V5Z 4H4, Canada
Received: 20-Oct-2020 Published: 10-Nov-2020
Abstract
In the work entitled “On the prediction of DNA-binding preferences of C2H2-ZF domains using structural models: application on human CTCF” we present a new computational method to predict the DNA-binding preferences for Cis2-His2 zinc finger (C2H2-ZF) protein-domains from their amino acid sequence or structure. The method uses the structures of protein-DNA complexes to calculate a set of knowledge-based statistical potentials. Given the low numbers of protein-DNA complexes with known structure, we supplement the set of structures with experimental yeast-one-hybrid interactions of more than 170000 sequence-designed C2H2-ZF domains. We have implemented a server to model the structure of any protein-DNA complex of this family and derive its theoretical Position Weight Matrix based on the best scores of interactions calculated with the statistical potentials. The approach is validated and applied to the human sequence of CTCF.
Keywords
Cis2-His2 Zinc-Finger transcription factors; Position weight matrix prediction; CTCF; Homology modeling of protein-DNA complexes; Protein-DNA statistical potentials
Description
Knowing the DNA-binding preferences of transcription factors (TFs) is foremost to understand gene regulation. Among TFs, Cis2- His2 zinc finger (C2H2-ZF) proteins are the largest family in higher metazoans [1] and in humans [2]. However, the DNA-binding preferences for many C2H2-ZF proteins are still unknown [2]. Since determining the DNA-binding preferences of TFs experimentally is both expensive and time-consuming [2,3], computational methods that complement experimental approaches can be very useful.
We use scoring functions based on structural and experimental data, called statistical potentials [4], to predict the DNA-binding preferences of C2H2-ZF proteins. Statistical potentials are scoring functions obtained from the analysis of a set of reference structures [5]. From this set of reference structures, we obtain frequencies of contacts between amino acids and dinucleotides. These frequencies are then used to calculate the statistical potentials by applying the inverse of the Boltzmann equation [6]. When analyzing the quality of protein structures, statistical potentials provide scores depending on how similar their contact frequencies are to the ones in the reference set [5]. In the case of C2H2-ZF, family we derive specific statistical potentials as follows: The reference set is constructed by collecting the structures of the members of the C2H2-ZF family complexed with DNA. Given the fact that the number of such structures is scarce and do not cover the entire spectrum of pairs of “amino acid” with “nucleotide” contacts, we complement the reference set with structural models of protein- DNA interactions from bacterial one-hybrid experiments [4,7]. The resulting statistical potentials are used to predict DNA-binding preferences of C2H2-ZF proteins from their amino acid sequence as described by means of a Position-Weight-Matrix (PWM) (Figure 1) [4]. We evaluate the performance of our approach by comparing the theoretical and experimental PWMs from bacterial one-hybrid experiments [7] and from the JASPAR database [8].
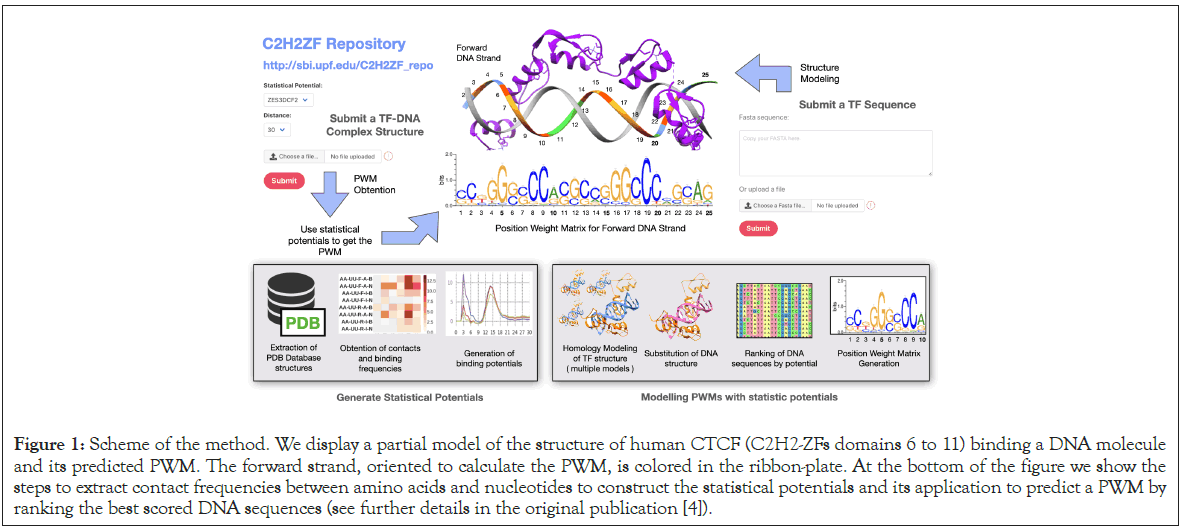
Figure 1. Scheme of the method. We display a partial model of the structure of human CTCF (C2H2-ZFs domains 6 to 11) binding a DNA molecule and its predicted PWM. The forward strand, oriented to calculate the PWM, is colored in the ribbon-plate. At the bottom of the figure we show the steps to extract contact frequencies between amino acids and nucleotides to construct the statistical potentials and its application to predict a PWM by ranking the best scored DNA sequences (see further details in the original publication [4]).
Finally, we apply our approach to predict the binding preferences of CTCF, a transcriptional repressor with a key role in genome compartmentalization [9]. Since no available CTCF-DNA complex structure covers the entire DNA-binding domain of CTCF, this results in incomplete theoretical PWM predictions. To overcome this limitation, we model almost the entire DNA-binding domain of CTCF: 10 out of 11 C2H2-ZF domains. The theoretical predictions are significantly similar to the canonical DNA-binding motif of CTCF between domains 4 to 8. Remaining domains correspond to other binding motifs upstream and downstream of the canonical binding domain [4], which has been described experimentally [10].
One strength of our method is that it provides more than one PWM per TF. Over 50% of predicted PWMs are significantly similar to their corresponding experimental PWM [4]. This means that, by scanning a DNA sequence with a set of PWMs of a TF, we can detect with higher reliability the binding site of a TF among the regions enriched by several matches of PWMs. Having several PWMs for one TF also implies that TFs might bind different DNA sequences using different interacting conformations. Zinc finger domains can interact with DNA in up to six different ways (or binding modes), each of which changes the orientation of the specificity residues of the finger with respect to the DNA and resulting motif [11]. An example illustrating this observation is CTCF, which displays different combinations of binding motifs spaced by a variable number of nucleotides (10). Our method is fast enough to be used in large collection of proteins containing C2H2- ZF domains. Out of all the eukaryotic C2H2-ZF proteins in the UNIPROT database [12], more than 70% of them can be analyzed by our method. The results of our study are available at: http:// sbi.upf.edu/C2H2ZF_repo. Besides the examples presented in the server, protein sequences containing the C2H2-ZF domains can be submitted to derived structural models and theoretical PWMs.
Acknowledgments
This work was supported by the Spanish Ministry of Economy (MINECO) and European Funds for Regional Development (EFRD) [BIO2017-85329-R][RYC-2015-17519]. BO, NFF and OF acknowledge the Council for the Catalan Republic. AM acknowledges a fellowship on Research Formation of “Generalitat de Catalunya” (FI). We acknowledge grant SGR17-1020 from Generalitat de Catalunya.
REFERENCES
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: Function, expression and evolution: Nat Rev Genet. 2009; 10(4):252-263.
- Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, et al. The human transcription factors. Cell. 2018; 172(4):650-665.
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014; 158(6):1431-1443.
- Meseguer A, Årman F, Fornes O, Molina-Fernández R, Bonet J, Fernandez-Fuentes N, et al. On the prediction of DNA-binding preferences of C2H2-ZF domains using structural models: Application on human CTCF. NAR Genom Bioinform. 2020; 2(3).
- Fornes O, Garcia-Garcia J, Bonet J, Oliva B. On the use of knowledge-based potentials for the evaluation of models of protein-protein, protein-DNA, and protein-RNA interactions. Adv Protein Chem Struct Biol. 2014; 94:77-120.
- Finkelstein AV, Badretdinov AYa, Gutin AM. Why do protein architectures have Boltzmann-like statisticals? Proteins. 1995; 23(2):142-150.
- Persikov AV, Wetzel JL, Rowland EF, Oakes BL, Xu DJ, Singh M, et al. A systematic survey of the Cys2His2 zinc finger DNA-binding landscape. Nucleic Acids Res. 2015; 43(3):1965-1984.
- Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020; 48(D1):D87-D92.
- Phillips JE, Corces VG. CTCF: Master weaver of the genome. Cell. 2009; 26;137(7):1194-1211.
- Nakahashi H, Kieffer Kwon K-R, Resch W, Vian L, Dose M, Stavreva D, et al. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 2013; 30;3(5):1678-1689.
- Garton M, Najafabadi HS, Schmitges FW, Radovani E, Hughes TR, Kim PM. A structural approach reveals how neighbouring C2H2 zinc fingers influence DNA binding specificity. Nucleic Acids Res. 2015; 30;43(19):9147-9157.
- UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019; 47(D1):D506-D515.
Citation: Meseguer A, Molina-Fernández R, Fernandez-Fuentes N, Fornes O, Oliva B (2020) Short Comment on: On the Prediction of DNA-binding Preferences of C2H2-ZF Domains Using Structural Models: Application on Human CTCF. J Data Mining Genomics Proteomics. 11:231. DOI: 10.35248/2153-0602.20.11.231.
Copyright: © 2020 Meseguer A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

