Indexed In
- Academic Journals Database
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- ResearchBible
- Ulrich's Periodicals Directory
- Electronic Journals Library
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Commentary - (2021) Volume 12, Issue 1
Short Commentary on: Comparative Proteomics of Mesembryanthemum crystallinum Guard Cells and Mesophyll Cells in the Transition from C3 to CAM
Qijie Guan1,2,3, Bowen Tan2, Jingkui Tian2,4* and Sixue Chen3,5*2College of Biomedical Engineering and Instrument Science, Zhejiang University, Hangzhou 310027, China
3Department of Biology, University of Florida, Gainesville 32610, USA
4Key Lab for Biomedical Engineering of Ministry of Education, Zhejiang-Malaysia Joint Research Center for Traditional Medicine, Zhejiang University, Hangzhou 310027, China
5Genetics Institute, Genetics Institute, Plant Molecular and Cellular Program, University of Florida, Gainesville 32610, USA
Received: 05-Jan-2021 Published: 26-Jan-2021, DOI: 10.35248/2153-0602.21.12.234
Abstract
In the work entitled “Comparative proteomics of Mesembryanthemum crystallinum guard cells and mesophyll cells in the transition from C3 to CAM”, we presented an interesting diurnal/diel proteomics analysis in M. crystallinum during its photosynthesis transition using the label-free method. We identified the proteins showing inverse responding patterns at each time point between the control (no transition) and treatment group (undergoing transition) in guard cells and mesophyll cells. This simple but useful method allowed us to focus on 165 and 151 proteins out of 1153 proteins in guard cells and mesophyll cells, respectively. The results facilitated understanding of plant photosynthesis plasticity at the single cell-type level.
Keywords
Mesembryanthemum crystallinum; Label-free proteomics; Diel pattern analysis; Inverse protein change pattern; C3 to CAM transition
Description
Although isobaric chemical labeling proteomics approaches like TMT and iTRAQ have better performance in multiplexing, they are quite costly and require extra experimental steps such as in vitro N-hydroxysuccinimide chemical reaction. In this study, we applied a label-free quantitative proteomics method [1,2]. Guard cells respond to complex interplay of signals, and guard cell proteomics may reveal interesting protein changes underlying the responses [3]. Computational methods can be useful in normalizing label-free proteomics data and using relative quantification to determine proteins involved in the biological processes. Here we developed a method to select and cluster proteins that exhibit temporal inverse abundances during the ice plant photosynthetic transition from C3 to Crassulacean Acid Metabolism (CAM).
A workflow designed to analyze inverse protein abundance patterns (Figure 1) was applied to M. crystallinum guard cells and mesophyll cells during the C3 to CAM transition induced by salt stress [4]. First, the raw data were analyzed using Proteome Discoverer 2.3 and the peak area for each protein was used for quantification. To analyze the changes of diel protein abundance in the two cell-types during the transition, median normalization followed by z-score transformation was used as a normalization method. Secondly, proteins have Person Correlation Coefficient (PCC) score less than -0.8 were selected as inversely expressed proteins [5] in the analysis of temporal changes of proteins during the C3-CAM transition (control versus treatment) in guard cells and mesophyll cells. Out of a total of 1153 identified proteins, 165 and 151 proteins showed inversed protein change patterns in guard cells and mesophyll cells, respectively. Thirdly, clustering of k-means based on Euclidean distance for control group proteins, in which each time point acts as a dimension, was used as the clustering method. Lastly, each cluster was analyzed using Gene Ontology (GO) enrichment [6] so that the functional aspects of the changed proteins in each cell type could be analyzed.
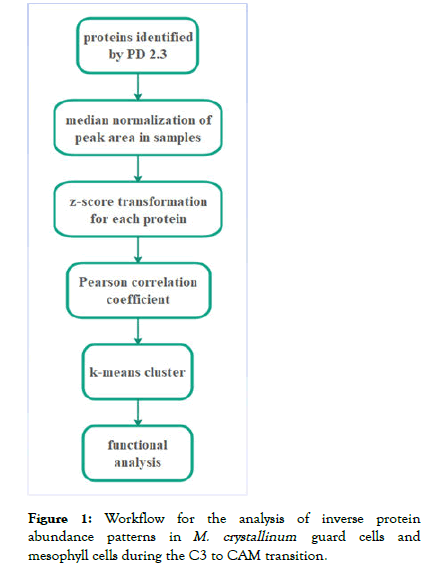
Figure 1: Workflow for the analysis of inverse protein abundance patterns in M. crystallinumguard cells and mesophyll cells during the C3 to CAM transition.
According to our proteomic result, the light response proteins in guard cells showed inversed abundance patterns during the transition, while those in mesophyll cells did not. This result is consistent with the inversed stomata movement behavior in the CAM plants [7]. Based on the protein changing patterns and the transition taking place before day 7 of treatment [8], we focused on cluster 3 of guard cell proteins and cluster 4 of mesophyll cell proteins. Another interesting finding from the enriched GO functions in two clusters is that ATP binding proteins and zinc ion binding proteins in two cell-types had very similar abundance changing patterns. This result indicates that there are common processes in the guard cells and mesophyll cells during the C3 to CAM transition.
Limitations
Our results highlight that the mechanisms underlying the shift from C3 to CAM in the two cell-types in M. crystallinum are different. A stringent cutoff of PCC was used to filter the proteins with the inversed patterns. However, some other potentially useful information may be sifted out. In addition, functional testing using reverse genetics in ice plant is challenging. Developing a model system (e.g., Kalanchoe blossfeldiana in Crassulaceae [9,10] for functional analysis of the candidate proteins in the C3 to CAM transition is needed.
Acknowledgements
This work was supported by a University of Florida Faculty Retention Fund, the China Scholarship Council (201706320126, 201506610004), the National Science and Technology Major Project of China (2019ZX09301004), National Science Foundation of China (81872973).
REFERENCES
- Li Z, Adams RM, Chourey K, Hurst G, Hettich R, Pan C. Systematic comparison of label-free, metabolic labeling, and isobaric chemical labeling for quantitative proteomics on LTQ Orbitrap Velos. J Proteome Res. 2012; 11(3):1582-1590.
- Latosinska A, Vougas K, Makridakis M, Klein J, Mullen W, Abbas M, et al. Comparative analysis of label-free and 8-Plex iTRAQ approach for quantitative tissue proteomic analysis. PloS One. 2015; 10(9):e0137048.
- Mach J. Guard cell proteome reveals signals and surprises. Plant Cell. 2008; 20(12):3185.
- Guan Q, Kong W, Zhu D, Zhu W, Dufresne C, Tian J. Comparative proteomics of Mesembryanthemum crystallinum guard cells and mesophyll cells in transition from C3 to CAM. J Proteomics. 2021; 231(1):104019.
- Chan YH. Biostatistics 104: Correlational analysis. Singapore Med J. 2003; 44(12):614-9.
- Ashburner M, Ball CA, Blake JA, David B, Butler H, Cherry JM, et al. Gene Ontology: Tool for the unification of biology. Nat Genet. 2000; 25(1): 25-9.
- Lawson T. Guard cell photosynthesis and stomatal function. New Phytologist. 2009; 181(1):13-34.
- Guan Q, Tan B, Kelley TM, Tian J, Chen S. Physiological changes in Mesembryanthemum crystallinum during the C3 to CAM transition induced by salt stress. Front Plant Sci. 2020; 11:283.
- Brulert J, Mricha A, Sossountzov L, Queiroz O. CAM induction by photoperiodism in green callus cultures from a CAM plant. Plant Cell Environ. 1987; 10(6) :443-449.
- Aida R, Shibata M. Transformation of Kalanchoe blossfeldiana mediated by Agrobacterium tumefaciens and transgene silencing. Plant Sci. 1996; 121(2):175-185.
Citation: Guan Q, Tan B, Tian J, Chen S (2021) Short Commentary on: Comparative Proteomics of Mesembryanthemum crystallinum Guard Cells and Mesophyll Cells in the Transition from C3 to CAM. J Data Mining Genomics Proteomics. 12:234.
Copyright: © 2021 Guan Q, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

