Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- CiteFactor
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research - (2019) Volume 10, Issue 5
Recent Developments in Genomic Selection for Minor Gene Quantitative Disease Resistance Plant Breeding
Dagnachew Bekele1,2*, Kassahun Tesfaye3 and Asnake Fikre42Ethiopian Institute of Agricultural Research, Addis Ababa, Ethiopia
3Ethiopian Biotechnology Institute, Addis Ababa, Ethiopia
4International Crop Research Institute for Semi-Arid Tropics, Addis Ababa, Ethiopia
Received: 23-Feb-2019 Published: 18-Apr-2019, DOI: 10.35248/2157-7471.10.478
Abstract
To speed up the development of improved crop varieties, genomics assisted plant breeding is becoming an important tool. With traditional breeding and marker assisted selection, there have been several achievements in breeding for diseases resistance. Most research for disease resistance has been focused on major disease resistance genes which are highly effective although very vulnerable to breakdown with rapid changes in pathogenic races. In contrast, breeding for minor gene quantitative resistance can produce more durable plant varieties although it is very slow and challenging breeding. As the genetic architecture of the plant disease resistance shifts from single major R genes to many minor quantitative genes, the most appropriate approach for molecular plant breeding is genomic selection (GS) than marker assisted selection or conventional breeding. With the advent of new genomic tools, GS has emerged as one of the most important approaches for predicting genotype performance to improve genetically complex quantitative traits. Consequently, GS helps to accelerate the rate of genetic gain in breeding by using whole genome sequence data to predict the breeding value of offspring. GS breeding for quantitative resistance will therefore necessitate whole genome prediction models and selection methodology as implemented for classical complex traits. With the implementation of GS for yield and other economically important traits, whole genome marker profiles are available for the entire set of breeding lines, enabling genomic selection for disease resistance with no additional direct cost. Therefore, recent developments in GS including a two stream GS + de novo GWAS models (GS+) and GS for combined highest level of quantitative resistance with R genes (QR +R gene) individuals are expected to further advance disease resistance plant breeding and briefly discussed.
Keywords
Breeding; Genomics selection; Diseases; Resistance; Quantitative resistance
Introduction
Conventional plant breeding strategies for selecting best and resistant plant genotypes for release depending on the phenotypic trait have proven to be of limited success. To develop improved varieties, any breeding program must have an efficient strategy for evaluating candidate genotypes for high yield, disease resistance, good agronomic performance, and better end use qualities [1]. Plant breeding for disease resistance varies depending on whether the resistance is considered to be qualitative or quantitative. The genetic architecture of disease resistance is closely tied to whether the resistance is quantitative or qualitative, and hence both the phenotypic and molecular breeding approaches must be matched accordingly. Qualitative disease resistance conditioned by single, major genes that does not have a complex genetic architecture. It is more suitable for identifying and mapping single resistance genes of large effect [2]. In contrast, quantitative disease resistance can be approached using whole genome prediction models developed for quantitative traits [2]. Genomic selection (GS) is an ideal approach for highly polygenic complex traits with lower heritability and a complex genetic architecture that are controlled by thousands of genes each with very small individual effects [3].
Recent advances in sequencing technologies helped to apply effective genotyping methods such as genotyping by sequencing (GBS) in different crop species with large number of markers for GS [1,2,4-6]. GS is highly advanced and accurate technique in estimating breeding values for complex quantitative traits compared to the traditional phenotypic selection or marker assisted selection (MAS). GS has more power to capture small effect loci that would be missed by MAS [7]. Besides, the use of dense genome wide markers increases the chance of markers being in linkage disequilibrium (LD) with QTL influencing the trait of interest [8], and determines to some extent how well genetic relationship and genetic architecture are captured by the genomic selection model [9].
In GS, breeding program representative individual are genotyped with dense, genome wide markers, such as SNPs and phenotyped for traits of interest. This set of individuals are called a training population which will be used to create a genomic prediction model [2,10-12]. This model estimates the sum of the additive genetic effects of the genome wide alleles on the trait of individuals and known as genomic estimated breeding values (GEBVs). In selection cycles, non phenotyped breeding materials are genotyped, with the same set of markers as training population. Then, appropriate prediction model is used to predict their GEBVs for the trait of interest. One of the most important features of GS is all selected markers need to be significant and used for prediction modeling [5-8].
Methods
Breeding for disease resistance
Breeding methods for disease resistance vary depending on whether the resistance is considered to be qualitative or quantitative. Qualitative plant disease resistance is resistance controlled by a single resistance (R) gene recognizing a virulence factors in a classic gene for gene mechanism. With molecular markers for R genes, direct selection for disease resistance can be implemented in the breeding programs. There has been significant effort on the identification of markers linked with major genes and mapping quantitative trait loci (QTLs) for disease resistance [2]. Several hundreds of R genes have been mapped across important crop plants, including rice [13,14], wheat [15-17], maize [18-20], soybean [21,22] and potato [23,24] as well as numerous other crop pathosystems [25,26].
Nevertheless, in applied plant breeding there are relatively very few examples of large scale implementation of MAS for disease resistance [2]. As reported by Miedaner & Korzun [2] in wheat and barley breeding, the lack of markers applied in commercial breeding for disease resistance could be due to having few diagnostic markers. Besides, few monogenic resistances are durable, and only a few QTLs with high effects have been successfully transferred into elite breeding material [27].
Currently marker assisted selection has failed significantly to improve polygenic trait [28,29]. Quantitative resistance is considered to be more durable than qualitative disease resistance. Unlike resistance based on R genes, quantitative resistance generally does not appear to be race specific [30,31]. With race specific resistance, the prevalent pathogen race is a component of the environment, leading to greater observed G × E interaction. By comparison, minor gene resistance that has no or minimal gene for gene interaction leads to much less G × E interaction. In breeding, yield stability is the ideal target, especially because good performance across years is desired. Stability in resistance, resistance with minimal G × E, is important for achieving yield stability, particularly in areas prone to epidemics.
Breeding for race nonspecific minor gene resistance is one way to minimize G × E of resistance. In addition to minimizing G × E, there are also quantitative genetic and genomic prediction models that can help improve breeding efficiency when G × E is present, as long as there is some genetic correlation between environments [29,32]. Therefore, as the genetic architecture of resistance shifts from single major R genes to a diffused architecture of many minor genes, the best approach for molecular plant breeding is shifting from marker assisted selection to genomic selection. With the implementation of GS for yield and other economically important traits, whole genome marker profiles are available for the entire set of breeding lines, enabling genomic selection for disease resistance with no additional direct cost.
Major steps and advantages of GS
Genomic selection showed great promise to strongly increase the rate of genetic improvement in plant breeding programs [8]. It allows a comparative larger gain from selection by estimating all marker effects simultaneously and subsequent selection of genetically superior individuals based on their genomic estimated breeding value (GEBV) [33], instead of using a few significant markers as in classical marker assisted selection.
As illustrated in Figure 1, GS uses a training population of individuals that have been both genotyped and phenotyped to develop a model that takes genotypic data from a candidate population of untested breeding materials and produces genomic estimated breeding values (GEBVs). These GEBVs say nothing of the function of the underlying genes but they are the ideal selection criterion. In the plant breeding context, untested breeding materials would belong to a broader population defined as a crop market class or the breeding program as a whole [1,11].
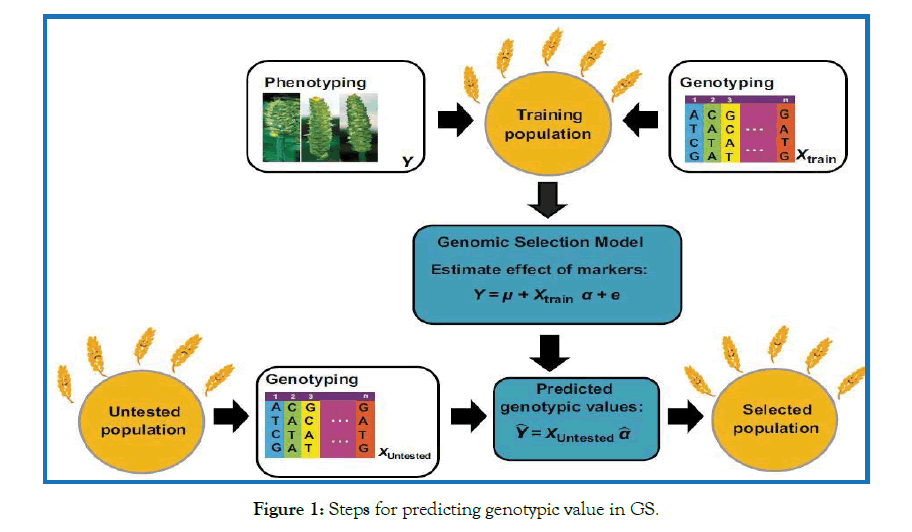
Figure 1: Steps for predicting genotypic value in GS.
Besides, very recently a two stream GS breeding scheme was developed [34] in which unutilized germplasm is systematically incorporated into a GS breeding pipeline again to test and predict the presence of new, highly effective allele combinations (Figure 2). In stream 1: Several pre-breeding materials with many favorable alleles from exotic germplasm are sequentially introduced into adapted germplasm. After many cycle of backcrossing and recombination to break linkages after the initial F1 cross between un-adapted and adapted material, selection of individuals from breeding population 1 is performed using a combination of GS + de novo GWAS models (GS+), in which the exotic QTL are fit as fixed effects and phenotype. The training population GS would be a subset of breeding population 1, that is, a fraction of breeding population 1 would be both genotyped and phenotyped, while the rest of breeding population 1 would be genotyped only.
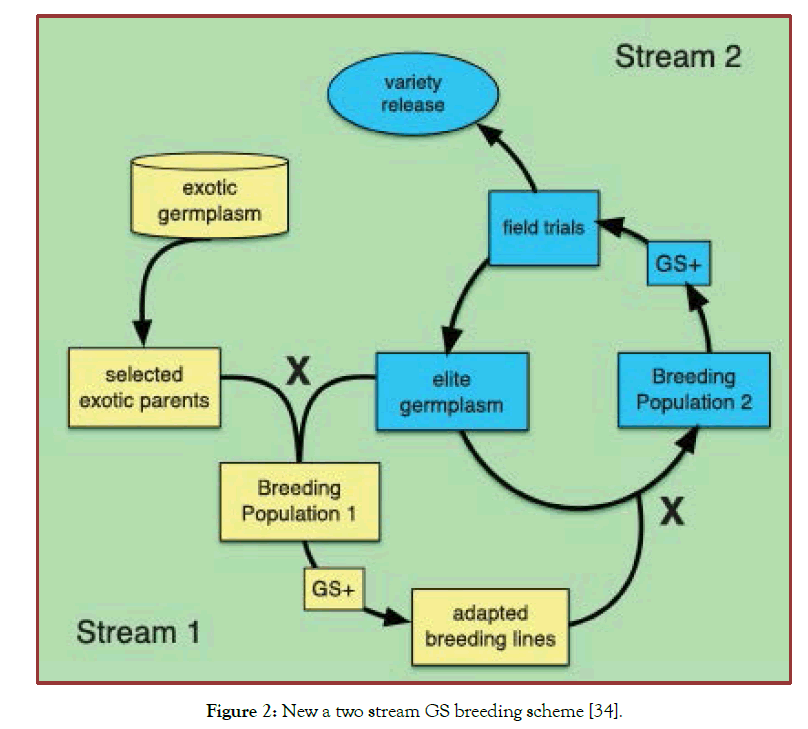
Figure 2: New a two stream GS breeding scheme [34].
Stream 2 continues the process of further refining and improving existing elite materials. Adapted materials from breeding population 1 are crossed into breeding population 2 where they are further refined using GS + de novo GWAS models, where the fixed effects would include valuable QTL identified based on GWAS performed in Breeding Population 2, the exotic QTL from Stream 1, or any other large effect QTL a breeder might normally target for trait improvement. Output from Stream 2 can be advanced toward variety release or fed back into stream 1 to serve as parents for further crossing and population development. This approach helps the breeders to learn directly from data on new and diverse germplasm and make rapid genetic gain.
The main factors that affect the accuracy of GS include the heritability of the trait, the rate of linkage disequilibrium decay, the marker density, and the number of individuals in the training population [9]. When LD decays more rapidly, then a greater number of markers and individuals for model training are needed. When predicting across populations, as in the case where previous breeding candidates are used for model training to predict new selection candidates, the relationship between the model training population and the selection candidates is important [35].
GS is a promising method for exploiting molecular genetic markers to design novel breeding programs and to develop new markers based models for genetic evaluation. In plant breeding, it provides opportunities to increase genetic gain of complex traits per unit time and cost. Therefore, for complex quantitative traits, GS provides higher selection accuracy in reduced time giving an accurate and good genetic gain per unit time as expected as compared with conventional phenotypic selection and MAS for complex traits. Besides, the fundamental difference between MAS and GS impacting the effectiveness of these two selection tools is scale [36]. MAS is limited in its ability to predict breeding values as it concentrates on a small number of QTLs that are tagged by markers with well-defined associations. In contrast, GS uses a dense set of markers from across the entire genome, assuring that all QTL are in LD with at least one SNP marker (Figure 3).
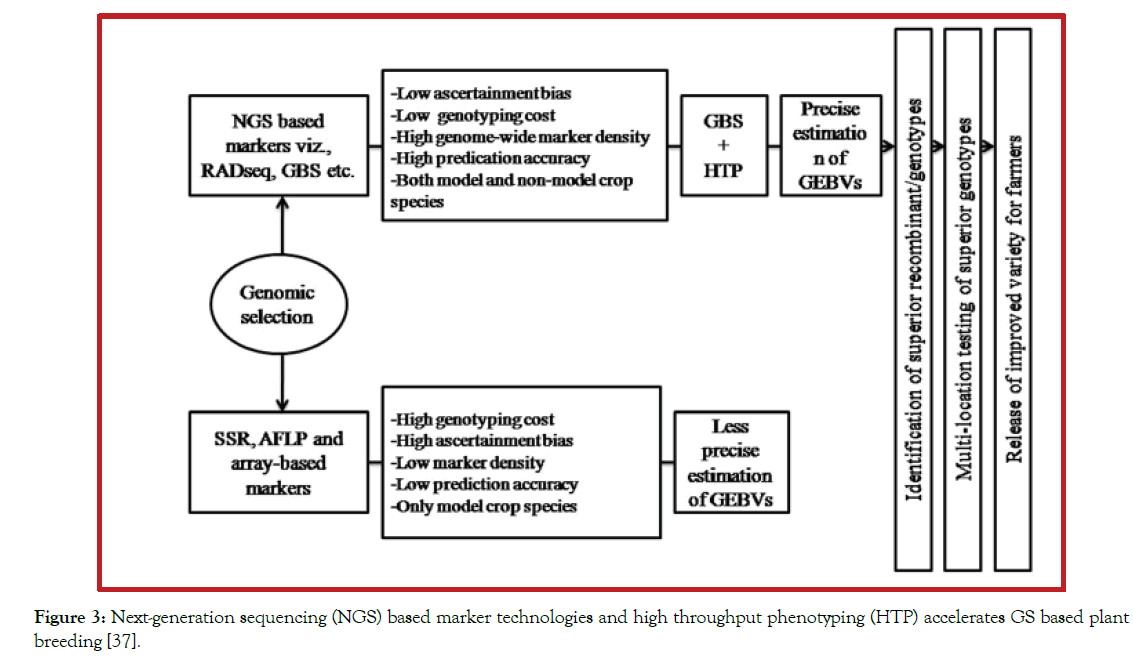
Figure 3: Next-generation sequencing (NGS) based marker technologies and high throughput phenotyping (HTP) accelerates GS based plant breeding [37].
Statistical models for GS
There are a variety of statistical models used to estimate breeding values in GS. Based on the crop species, trait considered, and breeding population design, the choice of statistical models used in GS has been shown to have a significant effect on prediction accuracy [37,38]. Very interestingly, for breeding crops like rice and wheat with large effect QTL, statistical models that incorporate a select number of molecular markers as fixed effects have been shown to contribute to improved prediction accuracy [34]. Two of the most commonly used models for purely quantitative traits are Genomic best linear unbiased prediction (G-BLUP) and ridge regression BLUP (RR-BLUP) [2]. G-BLUP is a mixed linear model, with individuals as random effects, and the covariance among individuals is assumed to be proportional to the genomic relationship matrix estimated with genome-wide markers.
G-BLUP is a modification of the conventional BLUP model [39], which uses pedigree relationships rather than genomic relationships. When using G-BLUP for prediction, both the model-training and validation individuals are included in the relationship matrix, but only the model training individuals have phenotypic data. RRBLUP is also a mixed linear model, but markers are considered random effects [40]. Covariance between markers is considered to be zero, and the marker variance is assumed to be the total genetic variance divided by the number of markers. This assumes that variance is equal for all markers, which enables many more marker effects to be estimated than there are phenotypic records [2]. Other suitable models for traits that fall between quantitative and qualitative inheritance are the Bayesian models Bayes-A, Bayes-B [8], Bayes-Cπ [41], and Bayesian LASSO [42]. With Bayes-A, each marker is assumed to have a unique variance. Bayes B is an extension of Bayes-A and allows some markers to have no effect.
Successes of GS in disease resistance breeding
In the past few years, there have been a number of research investigations that clearly demonstrated the application of many whole genome prediction models and GS approaches for disease resistance plant breeding (Table 1). These research efforts have confirmed the effectiveness of different GS models to capture and predict the genetic variation for disease resistance, particularly quantitative disease resistance [43-55].
| Crop | Disease type | References |
|---|---|---|
| Wheat | Rust | [43] |
| Wheat | Septoria tritici blotch | [44] |
| Barley | FHB* | [45] |
| Wheat | Stem rust | [46] |
| Wheat | FHB* | [44] |
| Wheat | FHB* | [47] |
| Wheat | Stem rust | [48] |
| Wheat | FHB* | [49] |
| Maize | NCLB** | [50] |
| Maize | Gibberella ear rot | [51] |
| Cassava | Cassava mosaic disease | [52] |
| Cassava | Cassava anthracnose disease | [52] |
| Barley | FHB* | [53] |
| Wheat | Stem rust | [54] |
| Wheat | Yellow rust | [54] |
| Wheat | FHB* | [55] |
Table 1: Major previous works demonstrating GS application for disease resistance breeding.
One of the most studied plant disease for the application of GS models in disease resistance plant breeding is rusts in wheat. The major rust pathogens of wheat are stem rust, yellow/stripe rust, and leaf brown rust [2]. Wheat rusts disease resistance can be either qualitative or quantitative, with several race specific R genes [56]. The R genes usually detected in the seedling stage. However, quantitative disease resistance loci generally confer resistance only in the adult plants, as a result it is referred as adult plant resistance (APR). APR has been shown to vary to some degree across environments, which could be due to race, temperature, or other unknown environmental factors.
Though disease resistance breeding efforts using major R genes for wheat rust have produced highly resistant varieties, their resistance breaks very shortly [2]. Therefore, breeding strategy for minor gene resistance highly preferred to generate varieties with durable disease resistance. Quantitative disease resistance is race nonspecific and sometimes effective against more than one rust species [57]. Previous studies on the application of GS for wheat rust disease resistance reported moderate to high prediction accuracies ranging from 0.3 to 0.8, and were able to predict both within and across environments with comparable accuracy [54,55]. Besides it is also indicated that GS can be relatively with small training population [46,48].
Besides, Fusarium head blight (FHB) is a serious disease in different parts of the world every year causing a very high yield loss and reduction of grain quality [58,59]. Different previous studies proven that resistance to FHB in wheat is quantitatively inherited [60,61] and that genetic variation for FHB resistance is predominantly additive [62,63] showing accumulation of resistance genes possible. The first report on GS models for FHB resistance in wheat found that various prediction models have high accuracy to be useful in breeding [55]. Besides, various studies evaluated RRBLUP as well as a nonlinear and a variable selection model on a diverse set of breeding lines for FHB and confirmed different traits associated with FHB resistance, could be predicted with moderate to high accuracy [47].
One of the major challenges with GS for disease resistance is that many disease resistances are highly heritable, which makes phenotypic selection hard to beat in both per-cycle and per unit genetic gain [46,47]. In order for GS to outperform phenotypic selection for quantitative disease resistance, it may be necessary to increase the selection intensity in addition to decreasing cycle time. For low-heritability traits, using genotype in addition to phenotype can substantially improve selection accuracy [64,65].
Recent advances in disease resistance breeding
The main advantage of using GS for durable disease resistance plant breeding is, quantitative resistance can be selected in the presence major R genes [2]. Several R genes confer a very high level of resistance and can severely mask the effect of other resistance QTLs in the background. In different plant breeding populations, both quantitative resistance loci and R genes are present together. In this case the ideal genotypes are those individuals with at least one effective R gene along with a high level of quantitative resistance. Therefore, this would lead to a very high level of disease resistance provided that the R gene is effective, and when the R gene is rendered ineffective, quantitative resistance in the background would still provide a good level of protection against yield loss.
The major advantage of genomics assisted plant breeding for disease resistance includes, it provides very good opportunities for combined selection of both R gene and quantitative resistance. The process mainly begins with crossing two parents that segregate for a race specific R gene based resistance to generate a population of selection candidates as illustrated in Figure 4. The selection candidates are then genotyped with WGS or GBS for genome wide markers. The selection candidates without effective R genes are then phenotyped for quantitative disease resistance. The phenotypes and genotypes are used to train genomic prediction model (GS model), which is then used to predict the level of quantitative resistance in the individuals with the effective R genes [2]. The individuals that combine the highest level of quantitative resistance with R genes (QR + R-gene candidates) are then selected for further advancement.
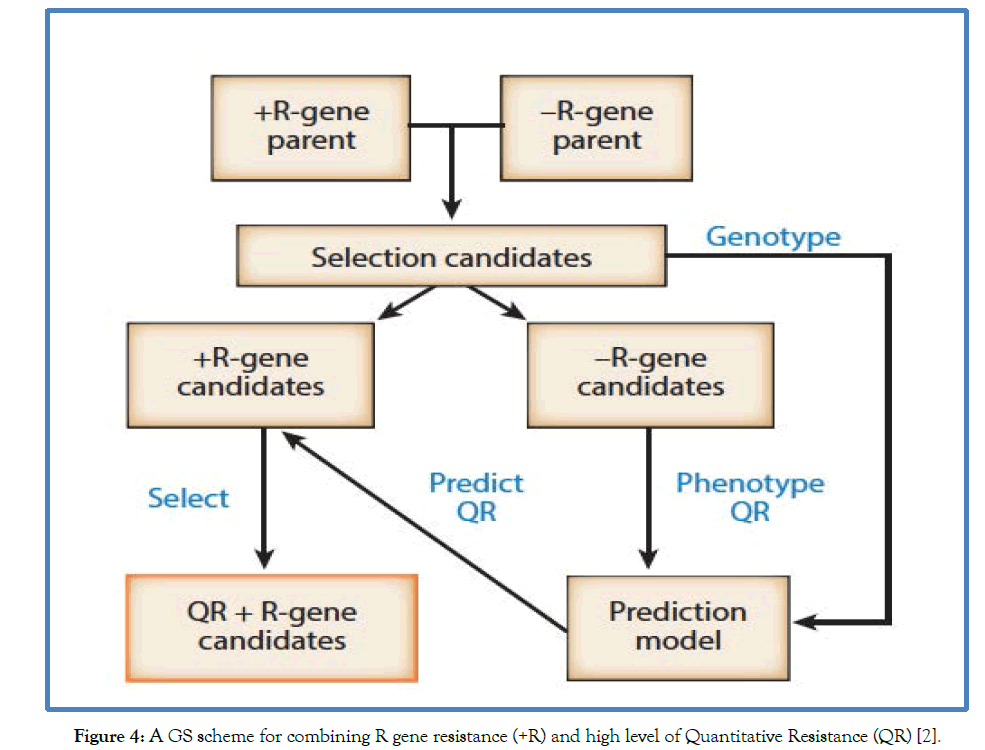
Figure 4: A GS scheme for combining R gene resistance (+R) and high level of Quantitative Resistance (QR) [2].
Results and Discussion
To speed up the development of improved crop varieties to feed an ever increasing world population, genomics assisted breeding is an important tool in any breeding program in the world. With traditional plant breeding and marker assisted selection, there has been several successes in breeding for diseases resistance. Most research works for disease resistance has been focused on major disease resistance genes which are highly effective although very vulnerable to breakdown with rapid changes in pathogenic races. There were several attempts in constructing genetic maps for important disease resistance genes and identifying several markers for MAS [2,10,11]. However, for many diseases, MAS quickly becomes very complex and intractable in an applied breeding program [2].
In contrast, breeding for minor gene quantitative resistance can produce more durable plant varieties although it is very slow and challenging breeding. To develop improved varieties with more durable disease resistance, the most appropriate approach is genomic selection (GS) than marker assisted selection or conventional breeding. The primary objective for implementing GS in plant breeding program is selection for high yield to reduce the time and costs associated with yield testing. As a result, GS is currently broadly accepted as an efficient method to improve genetically complex quantitative traits. Besides, recent developments in different sequencing technologies created excellent opportunities to apply different genotyping techniques including whole genome sequencing, genotyping by sequencing (GBS) in different crop species yielding an appropriate large number of markers for genomic. Therefore, GS helps to speed up the rate of genetic gain in breeding by using whole genome data to predict the breeding value of offspring. GS breeding for quantitative resistance will therefore necessitate whole genome prediction models and selection methodology as implemented for classical complex traits such as yield. With the application of GS for yield and other several economically important traits, whole genome marker profiles will be available for the entire set of breeding lines, enabling genomic selection for disease resistance with no additional direct cost.
GS has a great potential to improve plant breeding progress through increased selection intensity and decreased cycle time. In several studies, field based high throughput phenotyping highly demonstrated potential to measure different phenotypic traits faster and more accurately [66]. The combined power of genomics and phenomics is highly expected to lead to new eras in plant breeding and functional genomics [66,67]. Besides, current advances in disease resistance plant breeding through applications of a two stream GS breeding scheme using a combination of GS + de novo GWAS models (GS+) and genomic prediction model that combine the highest level of quantitative disease resistance with R genes (QR + R-gene candidates) are expected to further advance the implementation GS in disease resistance plant breeding.
Conclusion
Currently, there is a strong confidence and foundation for application of GS breeding for disease resistance plan breeding. Several studies successfully demonstrated the great potential of the current whole-genome prediction models efficiency to predict and select for quantitative disease resistance. With the implementation of GS for yield and other economically important traits, whole genome marker profiles will be available for the entire set of breeding lines, enabling genomic selection for disease resistance with no additional direct cost. Generally, genomic selection can increase breeding progress through increased selection intensity and decreased cycle time. Toward this major objective, the combined power of genomics and high throughput phenotyping technologies are expected to lead GS to new eras in disease resistance plant breeding and functional genomics studies. Besides, recent developments in disease resistance breeding, through a two stream GS breeding scheme using a combination of GS + de novo GWAS models (GS+) and GS model that combine the highest level of quantitative disease resistance with R genes (QR+R-gene) will further revolutionize the implementation GS in disease resistance plant breeding.
REFERENCES
- Chakradhar T, Hindu VReddy PSGenomic based breeding tools for tropical maize improvementGenetica 2017
- Poland J, Rutkoski JAdvances and challenges in genomic selection for disease resistanceAnn Rev Phytopathol 2016;54:79-98
- Poland JA, Balint Kurti PJ, WisserRJ, Pratt RC, Nelson RJShades of gray: The world of quantitative disease resistanceTrends Plant Sci 2009;14:21-29
- Hayes B, Bowman P, Chamberlain A, Verbyla K, Goddard MAccuracy of genomic breeding values in multi-breed dairy cattle populationsGenet Sel Evol 2009;41:51
- Varshney RKExciting journey of 10 years from genomes to fields and markets: some success stories of genomics assisted breeding in chickpea, pigeonpea and groundnutPlant Sci 2016;242:98-107
- Pandey MK, Roorkiwal M, Singh VK, Ramalingam A, Kudapa H, et alEmerging genomic tools for legume breeding: current status and future prospectsFront Plant Sci 2016;7:455
- Heffner EL, Sorrells ME, Jannink JLGenomic selection for crop improvementCrop Sci 2009;49:1-12
- Meuwissen THE, Hayes BJ, Goddard MEPrediction of total genetic value using genome-wide dense marker mapsGeneti 2001;157:1819-29
- Daetwyler HD, Pong-Wong R, Villanueva B, Woolliams JAThe impact of genetic architecture on genome-wide evaluation methodsGeneti 2010;185:1021-31
- Battenfield SD, Guzman C, Gaynor RC, Singh RP, Peña RJ, et alGenomic selection for processing and end-use quality traits in the CIMMYT spring bread wheat breeding programThe Plant Geno 2016
- Hickey JM, Chiurugwi T, Mackay I, Powell W, Eggen A, et alGenomic prediction unifies animal and plant breeding programs to form platforms for biological discoveryNat Genet 2017;49:1297-1303
- Bevan MW, Uauy C, Wulff BBH, Zhou J, Krasileva K, et alGenomic innovation for crop improvementNature 2017;543:346-354
- Song WY, Wang GL, Chen LL, Kim HS, Pi LYA receptor kinase-like protein encoded by the rice disease resistance gene, Xa21Science1995;270:1804-6
- Wang GL, Mackill DJ, Bonman JM, Mc Couch SR, Champoux MC, et alRFLP mapping of genes conferring complete and partial resistance to blast in a durably resistant rice cultivarGeneti 1994;136:1421-34
- Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino JA putative ABC transporter confers durable resistance to multiple fungal pathogens in wheatSci 2009;323:1360-63
- Periyannan S, Moore J, Ayliffe M, Bansal U, Wang XThe gene Sr33, an ortholog of barley mla genes, encodes resistance to wheat stem rust race Ug99Sci 2013;341:786-88
- Saintenac C, Zhang W, Salcedo A, Rouse MN, Trick HNIdentification of wheat gene Sr35 that confers resistance to Ug99 stem rust race groupScience 2013;341:783-86
- Kump KL, Bradbury PJ, Wisser RJ, Buckler ES, Belcher ARGenome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping populationNat Genet 2011;43:163-68
- Poland JA, Bradbury PJ, Buckler ES, Nelson RJGenome-wide nested association mapping of quantitative resistance to northern leaf blight in maizePNAS 2011;108:6893-98
- Ramakrishna W, Emberton J, Ogden M, San Miguel P, Bennetzen JLStructural analysis of the maize Rp1 complex reveals numerous sites and unexpected mechanisms of local rearrangementPlant Cell 2002;14:3213-23
- Bao Y, Vuong T, Meinhardt C, Tiffin P, Denny RPotential of association mapping and genomic selection to explore PI 88788 derived soybean cyst nematode resistancePlant Genom 2014
- Cregan PB, Mudge J, Fickus EW, Danesh D, Denny R, et alTwo simple sequence repeat markers to select for soybean cyst nematode resistance coditioned by the rhg1 locusTheor Appl Genet 1999;99:811-18
- 23Ballvora A, Ercolano MR, Weib J, Meksem K, Bormann CAThe R1 gene for potato resistance to late blight (Phytophthorainfestans) belongs to the leucine zipper/NBL/LRR class of plant resistance genesPlant J 2002;30:361-71
- Song J, Bradeen JM, Naess SK, Raasch JA, Wielgus SMGene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blightPNAS 2003;100:9128-33
- Ruffel S, Dussault MH, Palloix A, Moury B, Bendahmane AA natural recessive resistance gene against Potato virus Y in pepper corresponds to the eukaryotic initiation factor 4e (eIF4e)Plant J 2002;32:1067-75
- Zhou J, Loh YT, Bressan RA, Martin GBThe tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive responseCell 1995;83:925-35
- Miedaner T, Korzun VMarker-assisted selection for disease resistance in wheat and barley breedingPhytopatholo 2012;102:560-66
- Jannink J, Lorenz AJ, Iwata HGenomic selection in plant breeding: From theory to practiceBriefFunctGenom 2010;9:166-77
- 29Burgueno J, De Los Campos G, Weigel K, Crossa JGenomic prediction of breeding values when modeling genotype environment interaction using pedigree and dense molecular markersCrop Sci 2012;52:707-19
- Parlevliet JEDurability of resistance against fungal, bacterial and viral pathogens: Present situationEuphyti 2002;124:147-56
- Lindhout PThe perspectives of polygenic resistance in breeding for durable disease resistanceEuphytica 2002;124:217-26
- Kelly AM, Smith AB, Eccleston JA, Cullis BRThe accuracy of varietal selection using factor analytic models for multi-environment plant breeding trialsCrop Sci 47:1063-70
- BernardoRGenome wide selection when major genes are known.Crop Sci 2014;54:68-75
- Spindel JE, Begum H, Akdemir D, Collard B, Redona E, et alGenome-wide prediction models that incorporate de novo GWAS are a powerful new tool for tropical rice improvementHeredi 2016;116,395-408
- Pszczola M, Strabel T, Mulder H, Calus MReliability of direct genomic values for animals with different relationships within and to the reference populationJ Dairy Sci 2012;95:389-400
- Shikha M, Kanika A, Rao AR, Mallikarjuna MG, Gupta HS, et alGenomic selection for drought tolerance using genome-wide SNPs in maizeFrontPlant Sci 2017;8:550
- Bhat JA, Ali S, Salgotra RK, Mir ZA, Dutta S, et alGenomic selection in the era of next generation sequencing for complex traits in plant breedingFrontGenet 2016;7:221
- Daetwyler HD, Calus MP, Pong-Wong R, De Los Campos G, Hickey JMGenomic prediction in animals and plants: simulation of data, validation, reporting, and benchmarkingGenet 2013;193:347-365
- Henderson CRBest linear unbiased estimation and prediction under a selection modelBiometr 1975;31:423-47
- Whittaker JC, Thompson R, Denham MCMarker-assisted selection using ridge regressionGenet Res 2000;75:249-52
- Habier D, Fernando RL, Kizilkaya K, Garrick DJExtension of the Bayesian alphabet for genomic selectionBMC Bioinfo 2011;12:186
- Park T, Casella GThe Bayesian lassoJ Am Stat Assoc 2008;103:681-86
- Ornella L, Gonzalez-Camacho JM, Dreisigacker S, Crossa JApplications of genomic selection in breeding wheat for rust resistanceSambasivam Periyannan (ed), Wheat rust diseases: Methods and protocolsMeth Mol Bio 2017;1659
- Mirdita V, He S, Zhao Y, Korzun V, Bothe R, et alPotential and limits of whole genome prediction of resistance to Fusarium head blight and Septoria tritici blotch in a vast central European elite winter wheat populationTheor Appl Genet 2015;128:2471-81
- Sallam AH, Endelman JB, Jannink JL, Smith KPAssessing genomic selection prediction accuracy in a dynamic barley breeding populationPlant Geno 2015
- Rutkoski J, Singh RP, Huerta-Espino J, Bhavani S, Poland JGenetic gain from phenotypic and genomic selection for quantitative resistance to stem rust of wheatPlant Geno 2015
- Arruda, M, Brown, PJ, Lipka AE, Krill AM, Thurber C, et alGenomic selection for predicting head blight resistance in a wheat breeding programPlant Genom 2015
- Rutkoski JE, Poland JA, Singh RP, Huerta-Espino J, Bhavani SGenomic selection for quantitative adult plant stem rust resistance in wheatPlant Genom 2014
- Jiang Y, Zhao Y, Rodemann B, Plieske J, Kollers SPotential and limits to unravel the genetic architecture and predict the variation of Fusarium head blight resistance in European winter wheat (Triticum aestivum L)Heredi 2015;114:318-26
- Technow FB, Urger A, Melchinger AEGenomic prediction of northern corn leaf blight resistance in maize with combined or separated training sets for heterotic groupsG3 2013;3:197-203
- Riedelsheimer C, Endelman JB, Stange M, Sorrells ME, Jannink JLGenomic predictability of interconnected bi-parental maize populationsGeneti 2013);194:493-503
- Ly D, Hamblin M, Rabbi I, Melaku G, Bakare M, et alRelatedness and genotype x environment interaction affect prediction accuracies in genomic selection: a study in cassavaCrop Sci 2013;53:1312-25
- Lorenz AJ, Smith KP, Jannink JLPotential and optimization of genomic selection for Fusarium head blight resistance in six-row barleyCrop Sci 2012;52:1609-21
- Ornella L, Singh S, Perez P, Burgue J, Singh RGenomic prediction of genetic values for resistance to wheat rustsPlant Geno 2012;5:136-48
- Rutkoski J, Benson J, Jia Y, Brown-Guedira G, Jannink JLEvaluation of genomic prediction methods for Fusarium head blight resistance in wheatPlant Geno 2012;5:51-61
- Singh RP, Hodson DP, Jin Y, Lagudah ES, Ayliffe MAEmergence and spread of new races of wheat stem rust fungus: continued threat to food security and prospects of genetic controlPhytopatholol 2015;105:872-84
- William M, Singh R, Huerta-Espino J, Islas SO, Hoisington DMolecular marker mapping of leaf rust resistance gene Lr46 and its association with stripe rust resistance gene Yr29 in wheatPhytopatholo 2003;93:153-59
- Dexter J, Clear R, Preston KFusarium head blight: effect on the milling and baking of some Canadian wheatsCereal Chem 1996;73:695-701
- Pestka JJ, Smolinski ATDeoxynivalenol: Toxicology and potential effects on humansJ Toxicol Environ Health Part B 2005;8:39-69
- Liu S, Abate Z, McKendry AInheritance of Fusarium head blight resistance in the soft red winter wheat ErnieTheor Appl Genet 2005;110:454-61
- Van Ginkel M, Van Der Schaar W, Zhuping Y, Rajaram SInheritance of resistance to scab in two wheat cultivars from Brazil and ChinaPlant Dis 1996;80:863-67
- Bai GH, Shaner G, Ohm HInheritance of resistance to Fusarium graminearum in wheatTheor Appl Genet 2000;100:1-8
- Snijders CFusarium head blight and mycotoxin contamination of wheat: A reviewNeth J Plant Pathol 1990;96:187-98
- Heffner EL, Lorenz AJ, Jannink JL, Sorrells MEPlant breeding with genomic selection: gain per unit time and costCrop Sci 2010;50:1681-90
- Pszczola M, Veerkamp R, De Haas Y, Wall E, Strabel T, et alEffect of predictor traits on accuracy of genomic breeding values for feed intake based on a limited cow reference populationAnim 2013;7:1759-68
- Rutkoski J, Poland J, Mondal S, Autrique E, Parez LG, et alCanopy temperature and vegetation indices from high-throughput phenotyping improve accuracy of pedigree and genomic selection for grainYield in Wheat G3 2016;6
- Watson A, Ghosh S, Williams MJ, Cuddy WS, Simmonds J, et alSpeed breeding is a powerful tool to accelerate crop research and breedingNat Plants 2018;4:23-29.
Citation: Dagnachew B, Kassahun T, Asnake F (2019) Recent Developments in Genomic Selection for Minor Gene Quantitative Disease Resistance Plant Breeding. Plant Pathol Microbiol. 10: 478. doi: 10.35248/2157-7471.10.478
Copyright: © 2019 Dagnachew B, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

