Indexed In
- Academic Journals Database
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- ResearchBible
- Ulrich's Periodicals Directory
- Electronic Journals Library
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2023) Volume 14, Issue 4
In Silico Functional Prediction, and Expression Analysis in Response to Drought Stress of Natural Resistance-Associated Macrophage Protein (NRAMP) Gene Family in Maize
Md. Golam Rabby, Md. Mostafa Kamal, Md. Sakib Hasan, Md. Mossabbir Hossain and Md. Numan Islam*Received: 20-Jun-2023, Manuscript No. JDMGP-23-21908; Editor assigned: 23-Jun-2023, Pre QC No. JDMGP-23-21908 (PQ); Reviewed: 10-Jul-2023, QC No. JDMGP-23-21908; Revised: 17-Jul-2023, Manuscript No. JDMGP-23-21908 (R); Published: 25-Jul-2023, DOI: 10.4172/2153-0602.23.14.302
Abstract
Natural Resistance-Associated Macrophage Proteins (NRAMPs) function in plants as metal transition transporters to maintain metal homeostasis. The genome-wide identification and functional prediction of the maize NRAMPs have not yet been studied. The genome-wide analysis identified seven (ZmNRAMP1, ZmNRAMP2, ZmNRAMP3, ZmNRAMP4, ZmNRAMP5, ZmNRAMP6, ZmNRAMP7) proteins in maize with a molecular weight of 29.08 (ZmNRAMP2) to 63.20 (ZmNRAMP5) kDa. Except for ZmNRAMP1 and ZmNRAMP3, all ZmNRAMPs are essential proteins (isoelectric points greater than 7). The number of introns in ZmNRAMPs ranges from 1 to 13, which is almost consistent among the phylogenetic groups (A and B) between maize and Arabidopsis. All proteins showed multiple motifs (ranging from 6 to 10) that were consistent among the groups between maize and Arabidopsis. We also conducted transmembrane domains, cis-regulatory elements, chromosomal mapping, and gene ontology annotation that revealed significant functional attributes. Significantly increased ZmNRAMPs (ZmNRAMP1, ZmNRAMP2, and ZmNRAMP4) expression in vegetative and reproductive tissues may play an essential role in heavy metal absorption, transport, and homeostasis. During drought stress, significant upregulation of cc (ZmNRAMP1, ZmNRAMP2, ZmNRAMP4, and ZmNRAMP7) may play a role in drought tolerance or acclimatization in maize.
Keywords
NRAMP; Transporter; Drought; Gene expression; Maize
Introduction
Maize (Zea mays) is one of the world’s major food crops and is of substantial value to food, feed, pharmaceutical, and various industries [1,2]. It is widely used as a raw material in food product development and different bioenergy industries [3]. Maize is the world’s most productive and cultivated crop and a paradigm for genetics and genomics [4]. Among the top three crops in Asia, maize is one of them. In sub-Saharan Africa and Latin America, maize is considered the primary source of food security and economic development [5]. Various global environmental crises and stresses, including biotic and abiotic stresses, have posed a considerable threat to global maize production [6,7]. Therefore, it is necessary to clarify the molecular mechanism of maize in response to the unstable environmental situation [8]. Natural Resistance-Associated Macrophage Proteins (NRAMP) are known evolutionarily integral membrane proteins that play a crucial role in transporting a broad range of metal ions [9]. The NRAMPs are broadly distributed among various organisms, including bacteria, algae, yeast, plants, and animals [10]. The NRAMP1 gene of the NRAMP family was first identified in mice and has been recognized as a divalent metal transporter in plants, fungi, bacteria, insects, and mammals [11-13].
The NRAMPs transport iron and manganese to protect against bacterial infection [14]. The availability of NRAMPs among diverse species indicates the significance of their functions. Previous research has shown that NRAMP family proteins play an important role in structural stability and functional divergence across species. The NRAMP family proteins work as proton-coupled metal ion transporters and help transport Iron (Fe2+), Manganese (Mn2+), Zinc (Zn2+), Cadmium (Cd2+), Copper (Cu2+), Cobalt (Co2+), Nickel (Ni2+), and Aluminum (Al3+) [10,15-19]. These NRAMP family proteins have been implicated in the intracellular transport, translocation, uptake, and detoxification of transition metals [10]. The NRAMP transporter transports proton-coupled metal ions in different plants and plays a crucial role in plant growth and development, nutritional support, heavy metal balancing, and signal transduction [9]. Previous studies showed that the importance of the NRAMP family proteins in different plants had been investigated, including Arabidopsis thaliana [20], Populus alba [21], Oryza sativa [22], Theobroma cacao [23], Brassica napus [24], and Phaseolus vulgaris [25,26].
In Arabidopsis thaliana, AtNRAMP1 is found in the plasma membrane and works as an Iron (Fe2+), Manganese (Mn2+), and Cadmium (Cd2+) transporter [27,28], whereas AtNRAMP2 is found in the endomembrane and helps in the distribution of Manganese (Mn2+) between intracellular organelles [29,30]. The AtNARMP3 and AtNARMP4 transport Manganese (Mn2+) in plants that play a crucial role in photosynthesis and seed germination [31,32]. The AtNARMP5 has been identified as an Iron (Fe2+) and Cadmium (Cd2+) transporter [33]. The intracellular transportation of Cadmium (Cd2+) in plants is mediated by AtNARMP6 [27]. In rice, the NRAMP family proteins transport Iron (Fe), Manganese (Mn), Cadmium (Cd), and Arsenic (As) for the growth and development of rice. OsNARMP1 has been identified as a Fe, As, and Cd transporter [34-36], whereas OsNRAMP2 accumulates a high amount of Cadmium (Cd) in shoots [37]. The OsNRAMP3 helps transport Manganese (Mn) [38]. Furthermore, OsNRAMP4 transports Aluminum (Al) [16], whereas OsNRAMP5 transports Iron (Fe), Manganese (Mn), and Cadmium (Cd) [17,39]. OsNRAMP6 is found in the plasma membrane that works as an Iron (Fe), and Manganese (Mn) transporter [40].
However, the significant functions of many NRAMP transporters have been intensively investigated in Arabidopsis and other plants, but the NRAMP transporters in maize have not been characterized to date. Therefore, the current research was conducted for genome- wide identification and functional characterization of NRAMP transporters in maize by intensive bioinformatics investigations. In this study, we identified NRAMP genes from the maize genome and performed a comprehensive analysis of phylogeny, exon- intron structure, conserved motif, three-dimensional protein model, transmembrane domain architecture, and responsive motif cis-elements in the promoters, and chromosomal mapping. Furthermore, for a deep understanding of the functional roles of NRAMP transporters, the expression of different maize tissues under developmental and drought stress conditions, and gene ontology were also analyzed.
Materials and Methods
Identification of the ZmNRAMP gene family members in maize
The NRAMP proteins were identified following the Hidden Markov Model (HMM) profile of the conserved domains of AtNRAMP proteins based on the PFAM, CDD, and SMART databases. In brief, the complete maize genome (B73_RefGen_v4) sequence was downloaded from the maize database (http://www.maizesequence.org/index.html) and then TBtools software was used to construct a local database based on protein sequences. The known six AtNRAMP protein sequences were downloaded from TAIR (http://www.arabidopsis.org/) and used as queries to search against the maize genome via the local BLASTP program with an e-value of 1e-5 and where 50% identity was considered as the threshold. Further, all the NRAMP protein sequences of Arabidopsis have been aligned using ClustalX 2.1 (https://clustalx.software.informer.com/2.1/) for multiple sequence alignment, and the obtained alignments have been used to search the maize database using the online HMMER search tool. The results of HMMER search tools and BLASTP were compared and parsed by manual editing. For the confirmation of potential members of ZmNRAMP (PF01566) proteins, the PFAM database (http://pfam.xfam.org/), NCBI Conserved Domain Database (CDD) (https://www.ncbi.nlm.nih.gov/cdd/), and SMART database (http://smart.embl-heidelberg.de/) were used for removing redundant sequences. The remaining sequences were submitted to Inter-Pro-Scan (http://www.ebi.ac.uk/interpro/scan.html) to reconfirm the conserved domains. The maize genes were named according to the closeness of Arabidopsis genes. To analyze the physical and chemical parameters of the ZmNRAMP proteins, the ProtParam server (http://web.expasy.org/protparam/) was used [41-43].
Phylogenetic tree construction
The obtained protein sequence of ZmNRAMPs from the maize genome was aligned using ClustalX (version 2.1). Then, the phylogenetic tree of ZmNRAMP proteins was constructed by comparing AtNRAMP proteins following the Maximum Likelihood (ML) method and 1000 bootstrap replicates using MEGAX software [44,45].
Gene structures and conserved motifs analysis
The structure of ZmNRAMP proteins was investigated using the web-based GSDS server (http://gsds.cbi.pku.edu.cn/), and the conserved domains of the ZmNRAMP proteins were defined using Pfam (http://pfam.xfam.org/) and the TBtools software. The MEME suite (version 5.0.5) (http://meme-suite.org/tools/meme) was used to evaluate the conserved motifs and to identify the numbers of motifs based on the optimized E-values [45].
Prediction of three-dimensional modeling
The three-dimensional structures of representative ZmNRAMP proteins were constructed using the Phyre2 server (http://www.sbg.bio.ic.ac.uk/phyre2) to investigate structural differences and their effects on functions of the maize ZmNRAMP protein families [42].
Transmembrane domain prediction
For predicting transmembrane domains of the ZmNRAMP proteins, transmembrane topology crystal structures were constructed using the online tool HMMTOP (http://www.sacs.ucsf.edu/cgi-bin/hmmtop.py) and manually calculated the transmembrane helices [44].
Analysis of cis-regulatory elements in the promoters
The promoter sequences of ZmNRAMP genes were obtained from the maize whole-genome sequencing files. Subsequently, the upstream 2000 bp sequence relative to the translation initiation codon (ATG) of the promoter of each ZmNRAMP gene was selected as the promoter region, and the PlantCare online software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to predict the cis-acting elements in the promoters of ZmNRAMP genes. Finally, the prediction map of the ZmNRAMP gene’s promoters was visualized using TB tools [46].
The phenogram web server (http://visualization.ritchielab.org/phenograms/plot) was used to predict the chromosomal position of the ZmNRAMP genes. Phenogram creates ideograms with different colors according to the exact part of genes [47].
Expression analysis in different tissues and under drought stress conditions
To analyze the expression pattern of ZmNRAMP genes in different tissues [leaf, internodes, root, reproductive tissues (tassel, silk, and cob), embryo, endosperm, and seed], we downloaded the RNA-seq data of accession number SRP014652 (NCBI Bio-project 171684; PRJNA171684) from MaizeMine (http://maizemine.rnet.missouri.edu:8080/maizemine/begin.do), a freely accessible transcriptome database [48,49]. We also downloaded RNA-seq profiles of the accession number SRP347418 (NCBI Bio-project PRJNA782891: GEO accession GSE189392) from NCBI-SRA (https://www.ncbi.nlm.nih.gov/sra) to compare the expression patterns of respective transporter genes in control (drip irrigation) and under drought stress (negative pressure irrigation) [50]. An index of the maize genome sequence was built using bowtie2, and paired-end clean reads were mapped to the maize genome [51]. The cufflinks program was used to evaluate the expression level of the annotated genes in the reference genome [52]. The FPKM values were used to construct a heatmap using Tbtools [53].
Functional gene ontology analysis
To analyze the ZmNRAMP potential candidate genes and their corresponding IDs, they were subjected to the ShinyGO v0.61 database (http://bioinformatics.sdstate.edu/go/) to obtain Gene Ontology (GO) annotation against maize. GO enrichment was calculated by the p-value cut-off (FDR) at 0.05 for the genes [54].
Results
Identification and characterization of the maize NRAMP gene family
After confirming the sequences, a total of seven NRAMP genes were identified in the maize genome and named ZmNRAMP1, ZmNRAMP2, ZmNRAMP3, ZmNRAMP4, ZmNRAMP5, ZmNRAMP6, ZmNRAMP7. Table 1 shows the biochemical and physiological features of all proteins. The NRAMP genes in maize ranged from 265 (ZmNRAMP2) to 586 (ZmNRAMP5) amino acids. The molecular weight of the NRAMP proteins varies from 29.08 kDa (ZmNRAMP2) to 63.20 (ZmNRAMP5) kDa. Except for ZmNRAMP1 and ZmNRAMP3, all the NRAMP proteins have a pI value greater than 8, indicating that five of the NRAMP proteins are basic proteins. All proteins have positive GRAVY values, indicating that they are hydrophobic. In maize, four (ZmNRAMP4, ZmNRAMP5, ZmNRAMP6, and ZmNRAMP7) proteins are expected to be subcellular distributed in the plasma membrane, with three (ZmNRAMP1, ZmNRAMP2, and ZmNRAMP3) proteins in the vacuole.
| Transcript ID | Gene Name | Amino acid(aa) | Protein Molecular Weight(kDa) | pI | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|---|
| Zm00001d030846 | ZmNRAMP1 | 544 | 58.88 | 4.9 | 0.46 | Vacuole |
| Zm00001d048129 | ZmNRAMP2 | 265 | 29.08 | 9.05 | 0.52 | Vacuole |
| Zm00001d033367 | ZmNRAMP3 | 540 | 59.22 | 5.44 | 0.43 | Vacuole |
| Zm00001d014391 | ZmNRAMP4 | 550 | 59.52 | 8.26 | 0.57 | Plasma membrane |
| Zm00001d005479 | ZmNRAMP5 | 586 | 63.2 | 8.38 | 0.74 | Plasma membrane |
| Zm00001d015133 | ZmNRAMP6 | 551 | 58.94 | 8.01 | 0.58 | Plasma membrane |
| Zm00001d019327 | ZmNRAMP7 | 521 | 56.1 | 8.56 | 0.58 | Plasma membrane |
Table 1: Identification and characterization of NRAMP gene family in maize
Phylogenetic analysis of the NRAMP gene family in maize
According to the findings of phylogenetic analysis, seven ZmNRAMP genes have been classified into two subclasses (Figure 1). The two subclasses are named groups A and B, respectively. The results revealed that group-A includes 3 (ZmNRAMP1, ZmNRAMP2, and ZmNRAMP3) genes. Group-B includes 4 (ZmNRAMP4, ZmNRAMP5, ZmNRAMP6, and ZmNRAMP7) genes. The subclass group-A and B of the NRAMP gene families of maize may be functionally similar to the AtNRAMP2, AtNRAMP3, AtNRAMP4, AtNRAMP5 and AtNRAMP1, AtNRAMP16 genes in Arabidopsis, respectively.
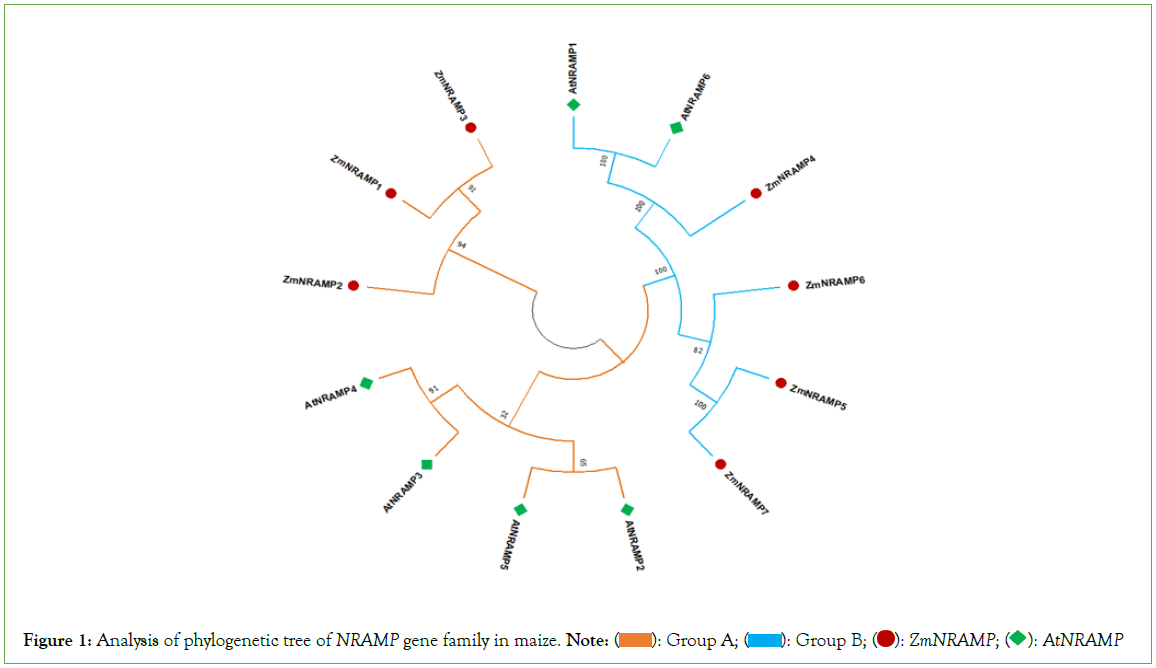
Figure 1: Analysis of phylogenetic tree of NRAMP gene family in maize. Note:  AtNRAMP
AtNRAMP
Analysis of the NRAMP gene family’s gene structures and conserved motifs in maize
The number of introns in the ZmNRAMP gene family ranges from 1 (ZmNRAMP2) to 13 (ZmNRAMP5) in various cluster groups (Figure 2). The group-A ZmNRAMP proteins (ZmNRAMP1, ZmNRAMP2, and ZmNRAMP3) have 1 to 3 introns. The group-B ZmNRAMP proteins (ZmNRAMP4, ZmNRAMP5, ZmNRAMP6, and ZmNRAMP7) have 9 to 13 introns. According to exon-intron structures and phylogenetic analysis, members of the same group have similar gene structures.
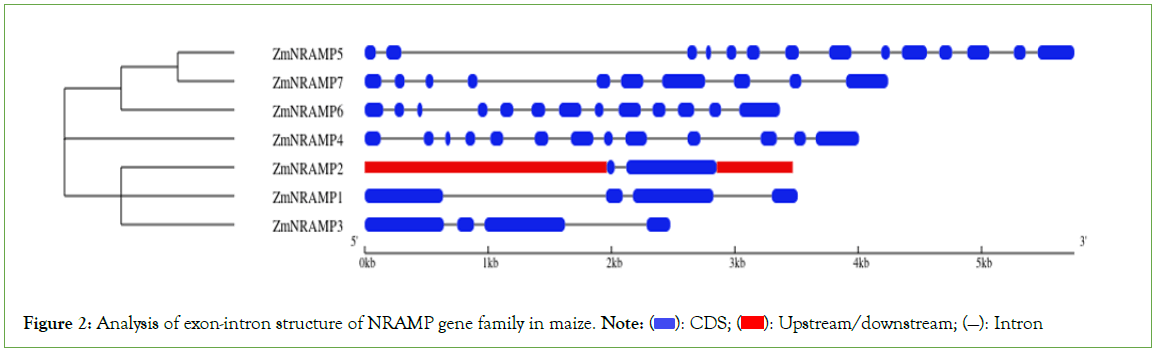
Figure 2: Analysis of exon-intron structure of NRAMP gene family in maize. Note:  Upstream/downstream; (—): Intron
Upstream/downstream; (—): Intron
Furthermore, a total of 10 motifs are identified and labeled as motifs 1 to 10 (Figure 3). The number of motifs in the ZmNRAMP family ranges from 6 (ZmNRAMP2) to 10 (ZmNRAMP4, ZmNRAMP5, ZmNRAMP6). Among all ZmNRAMP proteins, only ZmNRAMP4, ZmNRAMP5, and ZmNRAMP6 contain all 10 motifs. All the ZmNRAMP proteins have 6 (Motif 1, 3, 4, and 7, 8, 9) motifs. The group-A ZmNRAMP proteins (ZmNRAMP1, ZmNRAMP2, and ZmNRAMP3) have 6 to 9 motifs, and the group-B ZmNRAMP proteins (ZmNRAMP4, ZmNRAMP5, ZmNRAMP6, and ZmNRAMP7) have 9 to 10 motifs.
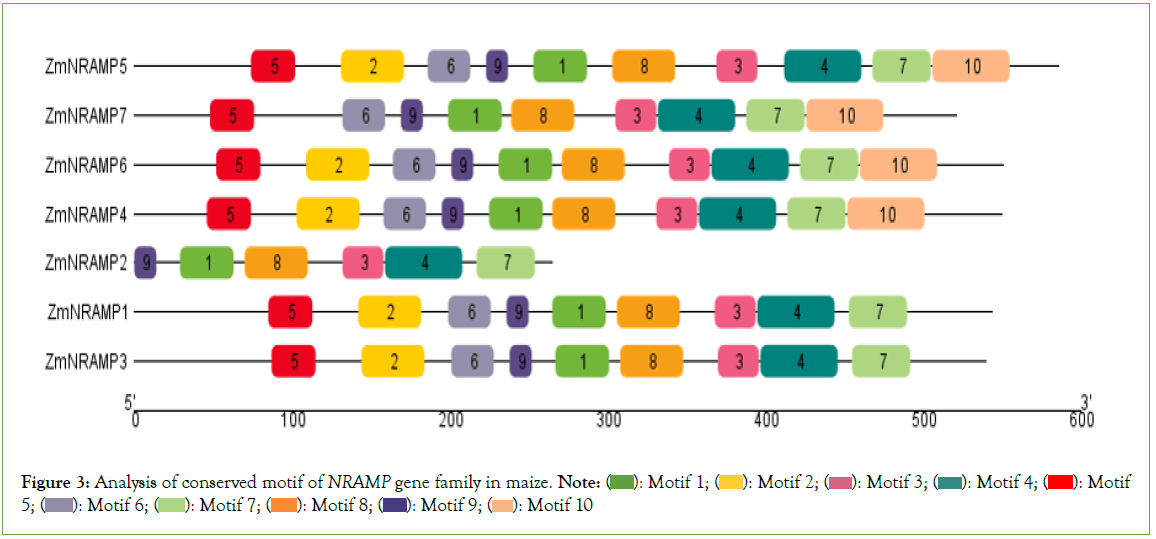
Figure 3: Analysis of conserved motif of NRAMP gene family in maize. Note: 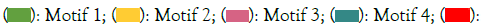 Motif 5;
Motif 5;  Motif 10
Motif 10
Analysis of three-dimensional modeling of NRAMP proteins in maize
The three-dimensional protein modeling reveals that all the ZmNRAMP proteins contain multiple alpha-helices, transmembrane helix, and coil structures (Figure 4). The percentage of the alpha- helix in the ZmNRAMP proteins varies from 69% (ZmNRAMP3 and ZmNRAMP7) to 80% (ZmNRAMP2). The percentage of transmembrane helices in the ZmNRAMP proteins varies from 48% (ZmNRAMP5) to 53% (ZmNRAMP3). All the ZmNRAMP proteins have a 100% confidence level (Table S1).
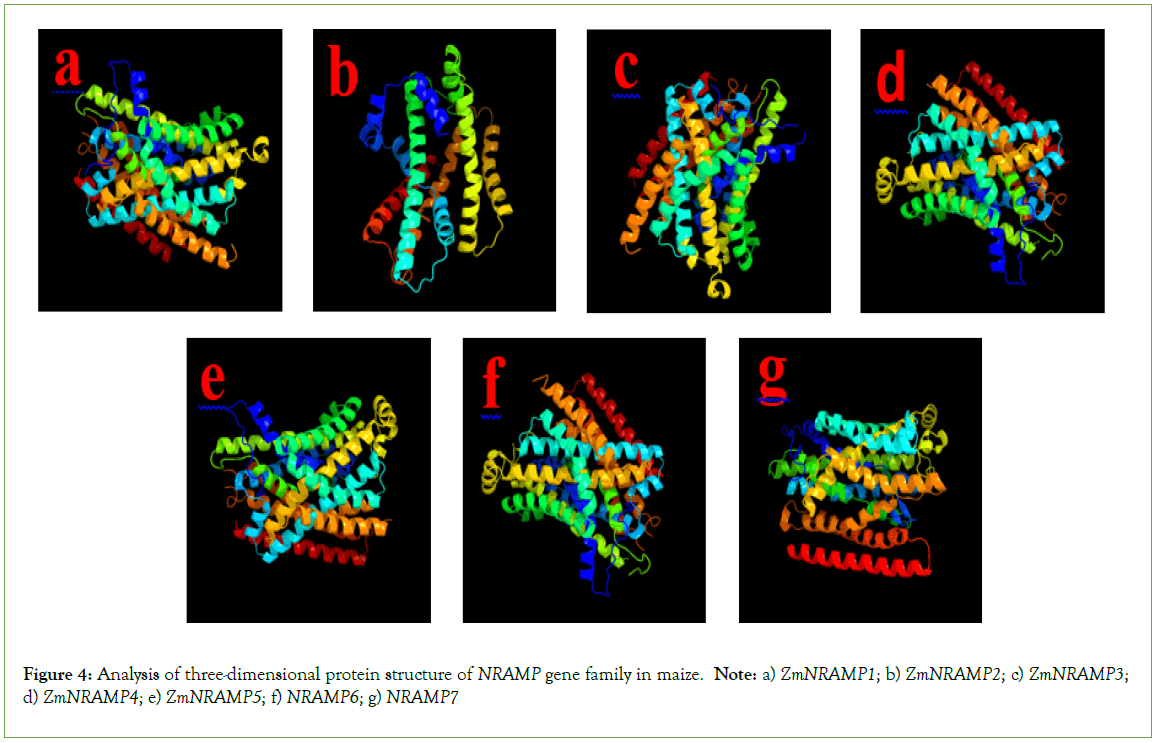
Figure 4: Analysis of three-dimensional protein structure of NRAMP gene family in maize. Note: a) ZmNRAMP1; b) ZmNRAMP2; c) ZmNRAMP3; d) ZmNRAMP4; e) ZmNRAMP5; f) NRAMP6; g) NRAMP7
domains of the NRAMP gene family in maize
As a consequence of the transmembrane domain analysis, multiple transmembrane helices were found in the ZmNRAMP gene family in maize. The ZmNRAMP proteins generally contain 1 to 12 transmembrane helices, and the intracellular sequences are extended from the N-terminus to the C-terminus (Figure 5). The four ZmNRAMPs (ZmNRAMP3, ZmNRAMP4, ZmNRAMP5, and ZmNRAMP6) contain all (12) the number of transmembrane helices. In the ZmNRAMP gene family, transmembrane domain length ranges from 265 (ZmNRAMP2) to 586 (ZmNRAMP5) and the number of transmembrane domains ranges from 6 to 12 (Table S2).
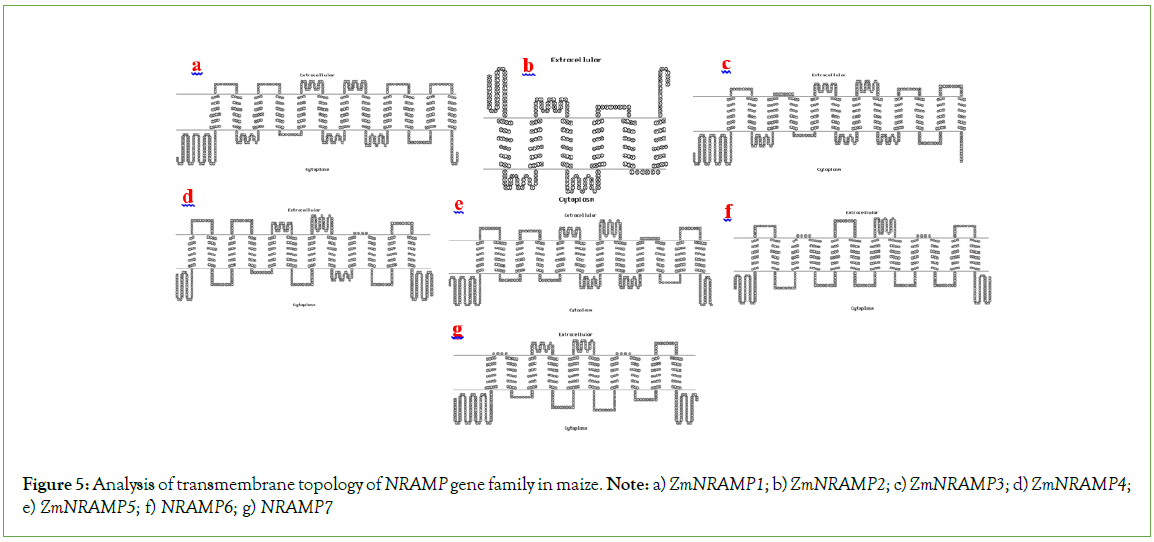
Figure 5: Analysis of transmembrane topology of NRAMP gene family in maize. Note: a) ZmNRAMP1; b) ZmNRAMP2; c) ZmNRAMP3; d) ZmNRAMP4; e) ZmNRAMP5; f) NRAMP6; g) NRAMP7
Analysis of cis-regulatory elements in the promoters of the NRAMP gene family in maize
The identification of putative cis-elements in promoter sequences is necessary for understanding gene regulation and function. To investigate the cis-elements of the ZmNRAMP genes, 2000-bp of the upstream sequence from the start codon was analyzed. The cis- elements of the ZmNRAMP genes were classified into three categories: plant growth and development, phytohormone responsiveness, and stress responses. The promoters of ZmNRAMP genes comprise three types of cis-elements. In the first category (related to growth and development), the elements included meristem expression (CAT-box), endosperm expression (GCN4_motif), seed-specific regulation (RY-element), and zein metabolism regulation (O2-site). In the second category, five types of phytohormone-responsive cis-elements were detected, auxin-responsive (TGA-element and AuxRE), Abscisic Acid-Responsive (ABRE), methyl jasmonate- responsive (CGTCA-motif and TGACG-motif), Gibberellin- Responsiveness (P-box, TATC-box, and GRE), and salicylic acid- responsive (TCA-element). In the third category (stress-responsive), the elements included MYB-binding sites involved in drought inducibility (MBS), Low-Temperature-Responsiveness (LTR), as well as defense and stress responsiveness (TC-rich repeats). All of the ZmNRAMP gene promoter sequences contained members of the three cis-element categories, with numbers ranging from 10 (ZmNRAMP4, ZmNRAMP5, ZmNRAMP6, and ZmNRAMP7) to 16 (ZmNRAMP1 and ZmNRAMP3), and the proportions of hormone-responsive elements were significantly greater. For example, ZmNRAMP1 and ZmNRAMP3 contained a total of 16 cis-elements. Among them, ZmNRAMP1 had 11 hormone-responsive cis-elements, and ZmNRAMP3 had 12 hormone-responsive cis- elements, (Figure 6, Table S3).
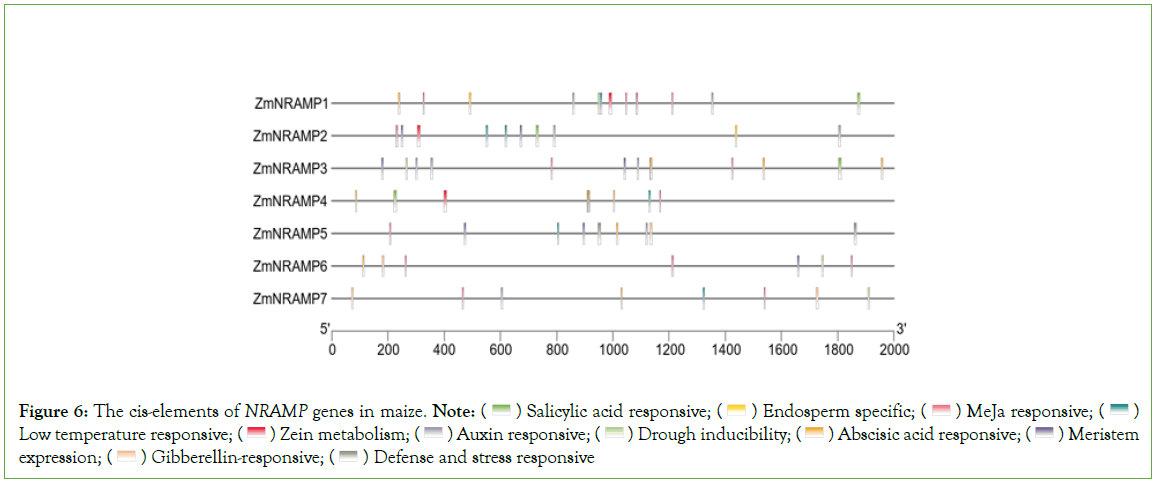
Figure 6: The cis-elements of NRAMP genes in maize. Note:  Low temperature responsive;
Low temperature responsive;  Meristem expression;
Meristem expression;  Defense and stress responsive
Defense and stress responsive
Chromosome mapping of maize NRAMP genes
The chromosomal mapping phenogram of seven ZmNRAMP genes revealed that all the genes were widely distributed on different chromosomes of maize. The phenogram detected the gene’s position from the chromosomal annotations. According to the combination of phylogenetic analysis and chromosomal mapping, chromosomes 1 and 9 contained the group A genes, and chromosomes 2, 5, and 7 included all the genes from group B. Chromosome 1 carried the ZmNRAMP1 and ZmNRAMP3 genes, chromosome 2 carried the ZmNRAMP5 gene, chromosome 5 carried the ZmNRAMP4 and ZmNRAMP6 genes, and chromosome 7 and 9 carried the ZmNRAMP7 and ZmNRAMP2 genes, respectively (Figure 7).
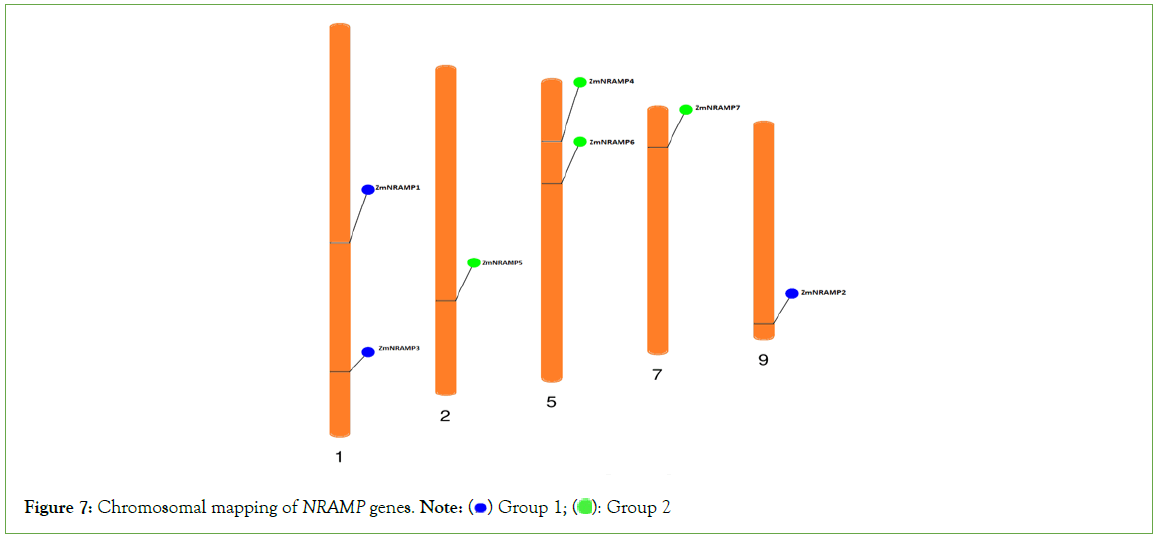
Figure 7: Chromosomal mapping of NRAMP genes. Note:  Group 2
Group 2
Expression analysis of maize NRAMP genes in different tissues and under drought stress conditions
The expression patterns of ZmNRAMPs were studied utilizing RNA-seq data acquired from specific tissues at various developmental stages from an online database. The normalized log2 (FPKM) value was displayed on the heat map (Figure 8). The ZmNRAMPs were classified into two categories based on their expression patterns. In the first cluster, three genes (ZmNRAMP1, ZmNRAMP2, and ZmNRAMP4) had a reasonably high expression level and were stable in practically all tissues at various developmental stages. In the second cluster, two genes, ZmNRAMP3 and ZmNRAMP5, were expressed at low levels in most tissues, except the leaf and anthers. The remaining two genes in the second cluster (ZmNRAMP6 and ZmNRAMP7) showed a wide range of expression patterns. Furthermore, some genes were highly expressed in multiple tissues, such as leaves, and internodes, roots, reproductive tissues, and seeds. For example, ZmNRAMP2 and ZmNRAMP4 were highly expressed in leaves, internodes, and roots and moderately expressed in seeds and reproductive tissues. On the other hand, the ZmNRAMP7 showed relatively high expression in leaves and roots (Figure 8A).
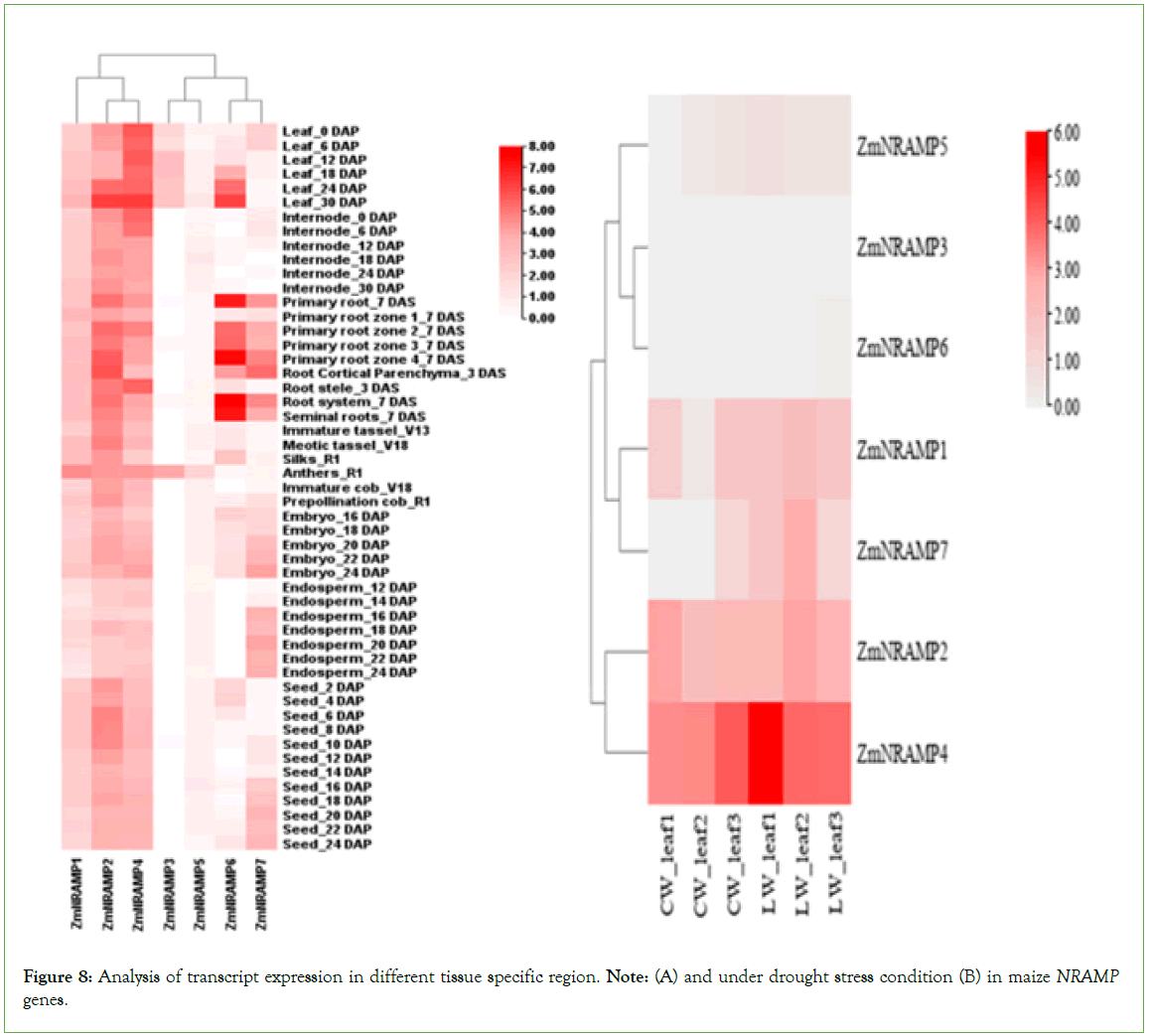
Figure 8: Analysis of transcript expression in different tissue specific region. Note: (A) and under drought stress condition (B) in maize NRAMP genes.
During drought stress, the ZmNRAMP1, ZmNRAMP2, ZmNRAMP4,
and ZmNRAMP7 genes were significantly upregulated in the leaves of maize. Among the ZmNRAMP family members, the transcript of the ZmNRAMP4 gene was highly upregulated, and ZmNRAMP1, ZmNRAMP2 and ZmNRAMP7 were moderately upregulated in the leaves of maize under drought stress, which may be due to their significant role in drought tolerance or acclimatization during drought stress in maize (Figure 8B).
Functional gene ontology analysis of maize NRAMP genes
The functional ontology of ZmNRAMP genes was predicted by the ShinyGO database, which showed the involvement of these genes in various molecular functions and biological processes. These genes showed significant molecular functions and biological processes, including cadmium, manganese, and aluminum transmembrane transporter activity; metal and inorganic cation transporter activity (Figure 9).
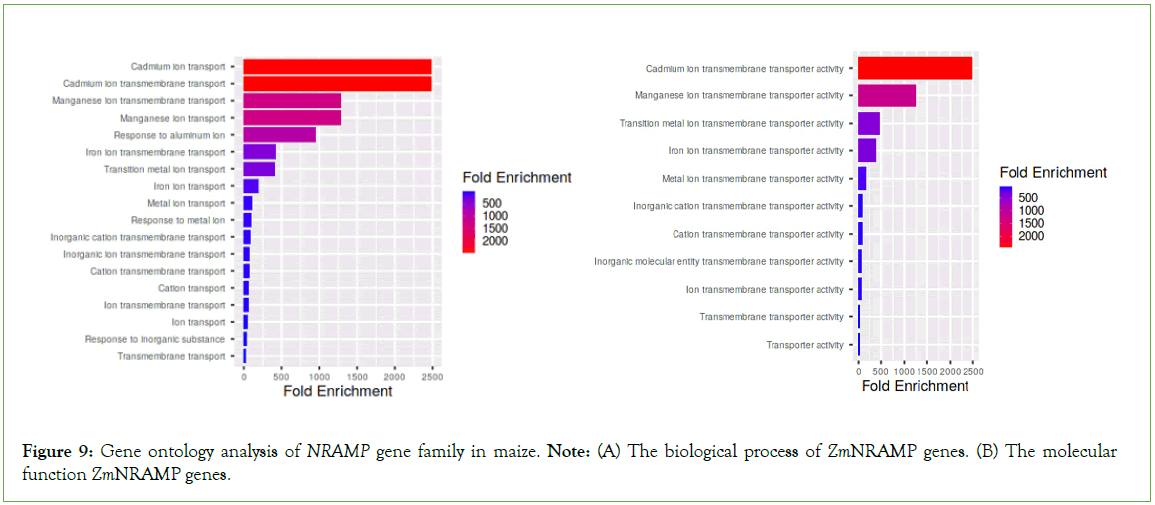
Figure 9: Gene ontology analysis of NRAMP gene family in maize. Note: (A) The biological process of ZmNRAMP genes. (B) The molecular function ZmNRAMP genes.
Discussion
Natural Resistance-Associated Macrophage Proteins (NRAMPs) are a prominent gene family in plants that play a significant role in the absorption and transport of heavy metals such as Iron (Fe), Copper (Cu), Zinc (Zn), Lead (Pb), Cadmium (Cd), and Manganese (Mn). The NRAMPs also show a crucial impact in response to heavy metal stress. There is an abundance of research that has been carried out on the NRAMP gene family and identified 48 members from 6 species, including Solanum tuberosum, Arabidopsis thaliana, Solanum Lycopersicum, Capsicum annuum, Oryza sativa, and Nicotiana attenuate [55]. In the current study, we identified ZmNRAMP family proteins from the maize genome database based on the conserved domain of identified AtNRAMP family proteins and assessed their functional and structural properties. The phylogenetic analysis of the current study indicated that the NRAMP gene families in maize and Arabidopsis are classified into two groups based on their identified protein sequences. They are named groups A and B. The exon-intron structure analysis of a specific plant genome indicates a close evolutionary similarity compared to another plant genome [56]. Exons and introns are integral structural components and play a significant impact on regulatory gene function [57]. Indeed, the structural variations of exon-introns, transmembrane domains, and motif compositions are significantly conserved within the same subfamilies [58]. However, functional diversity is the consequence of the structural changes of exons and introns [59]. The exon-intron structure analysis of our study revealed that ZmNRAMPs have 1 (ZmNRAMP2) to 13 (ZmNRAMP5) introns. Therefore, ZmNRAMP5 has the highest possibility of functional diversity, whereas ZmNRAMP2 has the least during maize genome evolution. The results of the conserved motif study, on the other hand, revealed that the number of motifs within sub-groups is nearly identical, and some amounts of motifs may have changed due to early evolution. The frequent mutations and deletions of conserved motifs are the result of functional alteration in plants [43]. In this current study, the gene structures and motif patterns of ZmNRAMPs are almost similar when compared to AtNRAMPs except for some variations. The consistent gene structures and motif patterns in maize and Arabidopsis genomes may share the same regulatory characteristics. However, some variation in gene structures and motif patterns among the members of ZmNRAMPs may be due to gene duplication and different evolutionary origins during the evolutionary process of maize genomes. The three-dimensional protein structures that consist of multiple alpha-helices reveal a protein’s evolutionary history and functional efficiency. The consistent transmembrane helices indicate that the relevant proteins are functionally active [43,48]. The analyzed three-dimensional protein structures of ZmNRAMP proteins contain multiple alphas and transmembrane helices. Consequently, the protein structures of ZmNRAMP and AtNRAMPs from different groups (A and B) are nearly identical, and this is the identification of functional activation.
The previous study suggested that the NRAMP proteins generally contained around 500 amino acid residues and 10 -12 transmembrane domains in different species. In rice, the OsNRAMP proteins contain 10-12 transmembrane domains and 518 -550 amino acid residues [60]. The PvNRAMP proteins in beans consist of 12 transmembrane domains and the amino acid length ranges from 507 to 554 [25]. Similarly, the CsNRAMP proteins in tea plants contain 3-12 transmembrane domains, and the length of amino acids ranged from 279 to 1373 [26]. However, in this current study, the identified ZmNRAMP proteins contain 6-12 transmembrane domains and the length of amino acids ranges from 265 to 586. The transmembrane domain structure analysis assesses the core membrane protein structure and overall topological characteristics. The impact of gene expressions and structural variations in plants was also evaluated through a transmembrane domain study [61]. According to the position of the N/C-terminal and the number of predicted transmembrane domains, the transmembrane domain structure could be divided into cytoplasmic, extracellular, and both cytoplasmic and extracellular N/C terminals. Among all the ZmNRAMPs, cytoplasmic N/C terminals were attributed to 10-12 TMs, which included ZmNRAMP1, ZmNRAMP3, ZmNRAMP4, ZmNRAMP5, ZmNRAMP6, ZmNRAMP7; only ZmNRAMP2 proteins were predicted in extracellular N/C terminals and showed 6 TMs; there were no both cytoplasmic and extracellular N/C terminal TMs. According to the phylogenetic tree analysis of maize and Arabidopsis NRAMP family members, they are phylogenetically related and may share common functional properties. Similarly, the comprehensive investigation into subcellular localization, exon-intron structure, conserved motifs, 3-D protein structure, a transmembrane domain, and cis-regulatory elements of promoter characteristics of maize and Arabidopsis showed similar structural properties. As a result, this current study estimated that the ZmNRAMP family proteins may have the same functional properties as Arabidopsis.
Our research has identified many growth and development, phytohormone, and stress-responsive cis-acting elements in the promoter region of ZmNRAMP genes. In addition, cis-acting components regulate the expression of stress-inducible genes, which helps them control significant biological processes [62]. The promoters of ZmNRAMP genes contain endosperm, seed, and meristem-specific cis-acting elements that lead to growth and developmental processes and control multiple stress signals. This is consistent with the essential functions of NRAMPs [63].
The expression patterns were performed to evaluate the probable functions of the ZmNRAMPs in maize growth and development. In contrast, ZmNRAMP genes were expressed at different levels in various tissues. Three genes (ZmNRAMP1, ZmNRAMP2, and ZmNRAMP4) in the ZmNRAMP gene family showed relatively high expression in leaves, internodes, and root zone. Indeed, ZmNRAMP2 from the first cluster was highly expressed in maize leaves and roots, and ZmNRAMP4 was highly expressed in maize leaves. Additionally, we have discovered the greater relative expression of ZmNRAMPs in leaves, internodes, and roots than in other maize tissues. Therefore, the greater expression patterns of ZmNRAMPs may play a significant role in the absorption, transport, and homeostasis of heavy metals. Interestingly, four ZmNRAMP genes (ZmNRAMP1, ZmNRAMP2, ZmNRAMP4, and ZmNRAMP7) were significantly upregulated in the leaves of maize during drought stress conditions. In particular, the ZmNRAMP4 gene was highly upregulated in maize leaves. Significant upregulation of transcripts of the above genes may be a consequence of their essential role in drought tolerance or acclimatization in maize. In general, the expression pattern of ZmNRAMPs in distinct tissues and drought stress showed their critical role in other organ development and controlling drought stress conditions in maize. This current study also identified the functional annotation of gene ontology, including molecular functions and biological processes. Gene ontology annotation revealed that ZmNRAMP proteins play a crucial role in cadmium, manganese, and aluminum transmembrane transporter activity. The ZmNRAMPs also play a role in metal and inorganic cation transporter activity.
Conclusion
In summary, this is the first compr`ehensive and systematic investigation of the NRAMP gene family in maize. We identified seven NRAMP genes from the maize genome and systematically investigated the genome-wide identification, chromosomal distribution, gene structure, conserved motifs, three-dimensional protein modeling, and transmembrane domain of NRAMP gene family members in maize. We subsequently constructed a phylogenetic tree. We also analyzed the cis-regulatory elements in the promoter region, chromosomal mapping, and gene ontology annotation of maize NRAMP genes. We further evaluated the expression patterns of NRAMP gene family members to determine their possible roles in the absorption and transport of heavy metals like Cd, Zn, Fe, and Mn in maize. Our research work will provide an inclusive framework for studying the maize NRAMP gene family. It will also contribute to the ZmNRAMP’s functional analysis and its applications.
Supplementary Materials
Supplementary Table S1. Characterization of three-dimensional modeling of the ZmNRAMP gene family in maize; Supplementary Table S2. Prediction of the transmembrane domain region of the ZmNRAMP gene family in maize; Supplementary Table S3. The numbers of different cis-elements in the ZmNRAMP genes.
Acknowledgments
This research work had been done in Advance laboratory of Nutrition and Food Technology department. We would like to thanks the Nutrition and Food Technology department.
Author´s Contributions
Md. Numan Islam and Md. Golam Rabby designed this study. Md. Golam Rabby and Md. Sakib Hasan performed comprehensive analyses. Md. Numan Islam, Md. Mostafa Kamal and Md. Mossabbir Hossain wrote and revised the manuscript. All authors read and approved the final manuscript.
Funding
This research received no external funding.
Data Availability
The readers interested in using the data may contact the corresponding author.
Declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
References
- Liu S, Wang X, Wang H, Xin H, Yang X, Yan J, et al. Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genet. 2013;9(9):e1003790.
[Crossref] [Google Scholar] [PubMed]
- Lu L, Hou Q, Wang L, Zhang T, Zhao W, Yan T, et al. Genome-wide identification and characterization of polygalacturonase gene family in maize (Zea mays L.). Int. J. Mol. Sci. 2021;22(19):10722.
[Crossref] [Google Scholar] [PubMed]
- Long T, Xu B, Hu Y, Wang Y, Mao C, Wang Y, et al. Genome-wide identification of ZmSnRK2 genes and functional analysis of ZmSnRK2.10 in ABA signaling pathway in maize (Zea mays L). BMC Plant Biol. 2021;21(1):309.
[Crossref] [Google Scholar] [PubMed]
- Hake S, Ross-Ibarra J. Genetic, evolutionary and plant breeding insights from the domestication of maize. eLife. 2015;4:e05861.
[Crossref] [Google Scholar] [PubMed]
- Cairns JE, Prasanna BM. Developing and deploying climate-resilient maize varieties in the developing world. Curr. Opin. Plant Biol. 2018;45:226-230.
[Crossref] [Google Scholar] [PubMed]
- Cairns JE, Sonder K, Zaidi PH, Verhulst N, Mahuku G, Babu R, et al. Maize production in a changing climate: Impacts, adaptation, and mitigation strategies. Adv. Agron. 2012;114:1-58.
- Fischer G. References and notes. Sov Oppos to Stalin. 2014;333:201–217.
- Hu W, Ren Q, Chen Y, Xu G, Qian Y. Genome-wide identification and analysis of WRKY gene family in maize provide insights into regulatory network in response to abiotic stresses. BMC Plant Biol. 2021;21:1-21.
[Crossref] [Google Scholar] [PubMed]
- Jin L, Enhe B, Zhuang Y, Hasi A, Gao F. Genome-wide identification and expression analysis of the NRAMP Family in melon. Int J Agric Biol. 2021;25(2):327-337.
- Nevo Y, Nelson N. The NRAMP family of metal-ion transporters. Biochim. Biophys. Acta-Mol. Cell Res. 2006;1763(7):609-620.
[Crossref] [Google Scholar] [PubMed]
- Vidal SM, Malo D, Vogan K, Skamene E, Gros P. Natural resistance to infection with intracellular parasites: Isolation of a candidate for Bcg. Cell J. 1993;73(3):469-485.
- Pinner E, Gruenheid S, Raymond M, Gros P. Functional complementation of the yeast divalent cation transporter family SMF by NRAMP2, a member of the mammalian natural resistance-associated macrophage protein family. J. Biol. Chem. 1997;272(46):28933-28938.
- Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, Boron WF, et al. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388(6641):482-488.
[Crossref] [Google Scholar] [PubMed]
- Supek F, Supekova L, Nelson H, Nelson N. A yeast manganese transporter related to the macrophage protein involved in conferring resistance to mycobacteria. Proc. Natl. Acad. Sci. U.S.A. 1996;93(10):5105-5110.
[Crossref] [Google Scholar] [PubMed]
- Colangelo EP, Guerinot ML. Put the metal to the petal: Metal uptake and transport throughout plants. Curr. Opin. Plant Biol. 2006;9(3):322-330.
[Crossref] [Google Scholar] [PubMed]
- Xia J, Yamaji N, Kasai T, Ma JF. Plasma membrane-localized transporter for aluminum in rice. Proc. Natl. Acad. Sci. U.S.A. 2010;107(43):18381-18385.
[Crossref] [Google Scholar] [PubMed]
- Sasaki A, Yamaji N, Yokosho K, Ma JF. Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. The Plant Cell. 2012;24(5):2155-2167.
[Crossref] [Google Scholar] [PubMed]
- Xiong H, Kobayashi T, Kakei Y, Senoura T, Nakazono M, Takahashi H, et al. AhNRAMP1 iron transporter is involved in iron acquisition in peanut. J. Exp. Bot. 2012;63(12):4437-4446.
[Crossref] [Google Scholar] [PubMed]
- Li JY, Liu J, Dong D, Jia X, McCouch SR, Kochian LV. Natural variation underlies alterations in Nramp aluminum transporter (NRAT1) expression and function that play a key role in rice aluminum tolerance. Proc. Natl. Acad. Sci. U.S.A. 2014;111(17):6503-6508.
[Crossref] [Google Scholar] [PubMed]
- Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, et al. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001;126(4):1646-1667.
[Crossref] [Google Scholar] [PubMed]
- Chen HM, Wang YM, Yang HL, Zeng QY, Liu YJ. NRAMP1 promotes iron uptake at the late stage of iron deficiency in polars. Tree Physiol. 2019;39(7):1235-1250.
[Crossref] [Google Scholar] [PubMed]
- Chang JD, Huang S, Yamaji N, Zhang W, Ma JF, Zhao FJ. OsNRAMP1 transporter contributes to cadmium and manganese uptake in rice. Plant Cell Environ. 2020;43(10):2476-2491.
[Crossref] [Google Scholar] [PubMed]
- Ullah I, Wang Y, Eide DJ, Dunwell JM. Evolution, and functional analysis of Natural Resistance-Associated Macrophage Proteins (NRAMPs) from theobroma cacao and their role in cadmium accumulation. Sci. Rep. 2018;8(1):14412.
[Crossref] [Google Scholar] [PubMed]
- Zhang XD, Meng JG, Zhao KX, Chen X, Yang ZM. Annotation and characterization of Cd-responsive metal transporter genes in rapeseed (Brassica napus). Biometals. 2018;31:107-121.
[Crossref] [Google Scholar] [PubMed]
- Ishida JK, Caldas DG, Oliveira LR, Frederici GC, Leite LM, Mui TS. Genome-wide characterization of the NRAMP gene family in Phaseolus vulgaris provides insights into functional implications during common bean development. Genet. Mol. Biol. 2018;41:820-833.
[Crossref] [Google Scholar] [PubMed]
- Li J, Duan Y, Han Z, Shang X, Zhang K, Zou Z, et al. Genome-wide identification and expression analysis of the NRAMP family genes in tea plant (Camellia sinensis). Plants. 2021;10(6):1055.
[Crossref] [Google Scholar] [PubMed]
- Cailliatte R, Schikora A, Briat JF, Mari S, Curie C. High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. The Plant Cell. 2010;22(3):904-917.
[Crossref] [Google Scholar] [PubMed]
- Nakanishi-Masuno T, Shitan N, Sugiyama A, Takanashi K, Inaba S, Kaneko S, et al. The crotalaria juncea metal transporter CjNRAMP1 has a high Fe uptake activity, even in an environment with high Cd contamination. Int. J. Phytoremediation. 2018;20(14):1427-1437.
[Crossref] [Google Scholar] [PubMed]
- Alejandro S, Cailliatte R, Alcon C, Dirick L, Domergue F, Correia D, et al. Intracellular distribution of manganese by the trans-Golgi network transporter NRAMP2 is critical for photosynthesis and cellular redox homeostasis. The Plant Cell. 2017;29(12):3068-3084.
[Crossref] [Google Scholar] [PubMed]
- Gao H, Xie W, Yang C, Xu J, Li J, Wang H, et al. NRAMP2, a trans‐Golgi network‐localized manganese transporter, is required for Arabidopsis root growth under manganese deficiency. New Phytol. 2018;217(1):179-193.
[Crossref] [Google Scholar] [PubMed]
- Lanquar V, Lelièvre F, Bolte S, Hamès C, Alcon C, Neumann D, et al. Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO J. 2005;24(23):4041-4051.
[Crossref] [Google Scholar] [PubMed]
- Lanquar V, Ramos MS, Lelièvre F, Barbier-Brygoo H, Krieger-Liszkay A, Krämer U, et al. Export of vacuolar manganese by AtNRAMP3 and AtNRAMP4 is required for optimal photosynthesis and growth under manganese deficiency. Plant Physiol. 2010;152(4):1986-1999.
[Crossref] [Google Scholar] [PubMed]
- Thomine S, Wang R, Ward JM, Crawford NM, Schroeder JI. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc. Natl. Acad. Sci. U.S.A. 2000;97(9):4991-4996.
[Crossref] [Google Scholar] [PubMed]
- Tiwari M, Sharma D, Dwivedi S, Singh M, Tripathi RD, Trivedi PK. Expression in Arabidopsis and cellular localization reveal involvement of rice NRAMP, OsNRAMP1, in arsenic transport and tolerance. Plant Cell Environ. 2014;37(1):140-152.
[Crossref] [Google Scholar] [PubMed]
- Curie C, Alonso JM, JEAN ML, Ecker JR, Briat JF. Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. J. Biochem. 2000;347(3):749-755.
[Crossref] [Google Scholar] [PubMed]
- Takahashi R, Ishimaru Y, Senoura T, Shimo H, Ishikawa S, Arao T, et al. The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. J. Exp. Bot. 2011;62(14):4843-4850.
[Crossref] [Google Scholar] [PubMed]
- Zhao J, Yang W, Zhang S, Yang T, Liu Q, Dong J, et al. Genome-wide association study and candidate gene analysis of rice cadmium accumulation in grain in a diverse rice collection. Rice. 2018;11(1):1-5.
[Crossref] [Google Scholar] [PubMed]
- Yang M, Zhang W, Dong H, Zhang Y, Lv K, Wang D, et al. OsNRAMP3 is a vascular bundles-specific manganese transporter that is responsible for manganese distribution in rice. PLoS One. 2013;8(12):e83990.
[Crossref] [Google Scholar] [PubMed]
- Ishikawa S, Ishimaru Y, Igura M, Kuramata M, Abe T, Senoura T, et al. Ion-beam irradiation, gene identification, and marker-assisted breeding in the development of low-cadmium rice. Proc. Natl. Acad. Sci. U.S.A. 2012;109(47):19166-19171.
[Crossref] [Google Scholar] [PubMed]
- Chen Y, Zhao X, Li G, Kumar S, Sun Z, Li Y, et al. Genome-wide identification of the Nramp gene family in Spirodela polyrhiza and expression analysis under cadmium stress. Int. J. Mol. Sci. 2021;22(12):6414.
[Crossref] [Google Scholar] [PubMed]
- Duan Y, Zhu X, Shen J, Xing H, Zou Z, Ma Y, et al. Genome-wide identification, characterization and expression analysis of the amino acid permease gene family in tea plants (Camellia sinensis). Genomics. 2020;112(4):2866-2874.
[Crossref] [Google Scholar] [PubMed]
- Rao KP, Richa TA, Kumar KU, Raghuram BA, Sinha AK. In silico analysis reveals 75 members of mitogen-activated protein kinase gene family in rice. DNA Res. 2010;17(3):139-153.
[Crossref] [Google Scholar] [PubMed]
- Wang C, Bao X, Li Y, Jiao C, Hou J, Zhang Q, et al. Data set for cloning and characterization of heterologous transporters in Saccharomyces cerevisiae and identification of important amino acids for xylose utilization. Data Br. 2015;4:119-126.
[Crossref] [Google Scholar] [PubMed]
- Wang C, Bao X, Li Y, Jiao C, Hou J, Zhang Q, et al. Cloning and characterization of heterologous transporters in Saccharomyces cerevisiae and identification of important amino acids for xylose utilization. Metab. Eng. 2015;30:79-88.
[Crossref] [Google Scholar] [PubMed]
- Yao X, Nie J, Bai R, Sui X. Amino acid transporters in plants: Identification and function. Plants. 2020;9(8):972.
[Crossref] [Google Scholar] [PubMed]
- Zeng D, Teixeira da Silva JA, Zhang M, Yu Z, Si C, Zhao C, et al. Genome-wide identification and analysis of the APETALA2 (AP2) transcription factor in Dendrobium officinale. Int. J. Mol. Sci. 2021;22(10):5221.
[Crossref] [Google Scholar] [PubMed]
- Zafar Z, Fatima S, Bhatti MF. Comprehensive analyses of NAC transcription factor family in almond (Prunus dulcis) and their differential gene expression during fruit development. Plants. 2021;10(10):2200.
[Crossref] [Google Scholar] [PubMed]
- Bishop EH, Kumar R, Luo F, Saski C, Sekhon RS. Genome-wide identification, expression profiling, and network analysis of AT-hook gene family in maize. Genomics. 2020;112(2):1233-1244.
[Crossref] [Google Scholar] [PubMed]
- Yang Y, Chai Y, Liu J, Zheng J, Zhao Z, Amo A, et al. Amino acid transporter (AAT) gene family in foxtail millet (Setaria italica L.): widespread family expansion, functional differentiation, roles in quality formation and response to abiotic stresses. BMC Genom. 2021;22(1):1-23.
[Crossref] [Google Scholar] [PubMed]
- Chen Z, Zhang Y, Zhang J, Fan B, Zhou Y, Cui X. Expression of AtAAP gene family and endosperm-specific expression of AtAAP1 gene promotes amino acid absorption in Arabidopsis thaliana and maize. J. Agron. 2021;11(8):1668.
- Hurtado-Guerrero R, Aalten DM. Structure of Saccharomyces cerevisiae chitinase 1 and screening-based discovery of potent inhibitors. J. Chem. Biol. 2007;14(5):589-599.
[Google Scholar] [Crossref] [PubMed]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7(3):562-578.
[Crossref] [Google Scholar] [PubMed]
- Han Y, Li X, Cheng L, Liu Y, Wang H, Ke D, et al. Genome-wide analysis of soybean JmjC domain-containing proteins suggests evolutionary conservation following whole-genome duplication. Front. Plant Sci. 2016;7:1800.
[Crossref] [Google Scholar] [PubMed]
- Jeyasri R, Muthuramalingam P, Satish L, Adarshan S, Lakshmi MA, Pandian SK, et al. The role of OsWRKY genes in rice when faced with single and multiple abiotic stresses. J. Agron. 2021;11(7):1301.
- Tian W, He G, Qin L, Li D, Meng L, Huang Y, et al. Genome-wide analysis of the NRAMP gene family in potato (Solanum tuberosum): Identification, expression analysis and response to five heavy metals stress. Ecotoxicol. Environ. Saf. 2021;208:111661.
[Crossref] [Google Scholar] [PubMed]
- Pan X, Hu M, Wang Z, Guan L, Jiang X, Bai W, et al. Identification, systematic evolution and expression analyses of the AAAP gene family in Capsicum annuum. BMC Genom. 2021;22(1):463.
[Crossref] [Google Scholar] [PubMed]
- Dong C, Li F, Yang T, Feng L, Zhang S, Li F, et al. Theanine transporters identified in tea plants (Camellia sinensis L.). Plant J. 2020;101(1):57-70.
[Crossref] [Google Scholar] [PubMed]
- Zhang C, Kong N, Cao M, Wang D, Chen Y, Chen Q. Evolutionary significance of amino acid permease transporters in 17 plants from Chlorophyta to Angiospermae. BMC Genom. 2020;21(1):1-1.
[Crossref] [Google Scholar] [PubMed]
- Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001;305(3):567-580.
[Crossref] [Google Scholar] [PubMed]
- Mani A, Sankaranarayanan K. In silico analysis of natural resistance-associated macrophage protein (NRAMP) family of transporters in rice. Protein J. 2018;37:237-247.
[Crossref] [Google Scholar] [PubMed]
- Zhao H, Ma H, Yu L, Wang X, Zhao J. Genome-wide survey and expression analysis of amino acid transporter gene family in rice (Oryza sativa L.). PloS one. 2012;7(11):e49210.
[Crossref] [Google Scholar] [PubMed]
- Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters. Trends Plant Sci. 2005;10(2):88-94.
[Crossref] [Google Scholar] [PubMed]
- Wan Y, King R, Mitchell RA, Hassani-Pak K, Hawkesford MJ. Spatiotemporal expression patterns of wheat amino acid transporters reveal their putative roles in nitrogen transport and responses to abiotic stress. Sci. Rep. 2017;7(1):5461.
[Crossref] [Google Scholar] [PubMed]
Citation: Rabby Md. G, Kamal Md. M, Hasan Md. S, Hossain Md. M, Islam Md. N (2023) In Silico Functional Prediction, and Expression Analysis in Response to Drought Stress of Natural Resistance-Associated Macrophage Protein (NRAMP) Gene Family in Maize. J Data Mining Genomics Proteomics. 14:302.
Copyright: © 2023 Rabby Md. G, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

