Indexed In
- Open J Gate
- Genamics JournalSeek
- ResearchBible
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- Publons
- MIAR
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research - (2023) Volume 12, Issue 6
Herbicidal Quassinoids Isolated from Ailanthus altissima Leaves
Young Sook Kim1, Surk-Sik Moon2* and Jung Sup Choi1*2Department of Chemistry, Kongju National University, Gongju 314-701, Republic of Korea
Received: 16-Nov-2023, Manuscript No. BABCR-23-23962; Editor assigned: 20-Nov-2023, Pre QC No. BABCR-23-23962 (PQ); Reviewed: 04-Dec-2023, QC No. BABCR-23-23962; Revised: 11-Dec-2023, Manuscript No. BABCR-23-23962 (R); Published: 18-Dec-2023, DOI: 10.35248/2161- 1009.23.12.517
Abstract
Bioassay-guided fractionation of the methanolic extract of Ailanthus altissima leaves led to the isolation of one new quassinoid, named 6-α-tigloyloxyailanthone (3), and three known quassinoids, ailanthone (1), 13, 18-dehydroglaucarubinone (2), and 6-α-tigloyloxychaparrinone (4) by a series of chromatographic methods. The structures of the isolates were established by one-dimensional (1D) and 2D-Nuclear Magnetic Resonance (NMR) analysis along with High- Resolution Time-Of-Flight Mass Spectrometry (HRTOFMS) and chemical methods. In addition, herbicidal activity against five grassy weeds and five broad-leaf weeds was evaluated.
Keywords
Quassinoids; Ailanthone; Ailanthus altissima; Simaroubaceae; Herbicidal activity
Abbreviations
COSY: Correlation Spectroscop Y; HMBC: Heteronuclear Multiple Bond Correlation; HPLC: High-Performance Liquid Chromatography; HRTOFMS: High-Resolution Time-Of-Flight Mass Spectrometry; NMR: Nuclear Magnetic Resonance; NOE: Nuclear Overhauser Effect; HSQC: Heteronuclear Single Quantum Correlation; ROESY: Rotating-Frame Nuclear Overhauser Effect Spectroscopy
Introduction
Quassinoids are a group of degraded triterpenes found in the family Simaroubaceae that undergo extensive oxidative biodegradation, leaving their carbon skeleton highly oxygenated [1-4]. Quassinoids are classified into five groups according to their basic structure, C-18, C-19, C-20, C-22, and C-25. The C-20 quassinoids have been extensively investigated due to their antileukemic activity discovered in the early 1970s [5]. Since 1960, hundreds of quassinoids have been isolated and identified from plants, but they seem harder to synthesize due to the presence of their highly oxygenated carbon framework. Many quassinoids display a wide range of biological activities in vitro or in vivo, including antitumor [6], antimalarial [7], antiviral [8], anti-inflammatory [9], antifeedant [10], insecticidal [11], antiulcer [12], and herbicidal activities [13]. In our ongoing investigations of structurally unique bioactive agents, such as herbicides derived from traditional plants, systematic phytochemical studies of Ailanthus altissima resulted in the isolation of one new quassinoid (3) and three known quassinoids (1, 2, and 4). In this paper, we describe the isolation, structural elucidation, and evaluation of the herbicidal activities of all isolates obtained.
Materials and Methods
Experimental details
Plant material: Seeds of 10 weed species (S. bicolor, E. crus-galli, A. smithii, D. sanguiinalis, P. dichotomiflorum, S. nigrum, A. indica, A. avicennae, X. strumarium, C. japonica) were germinated in flats in a commercial greenhouse substrate and watered with tap water. The weeds were grown in a greenhouse at 30 ± 3/20 ± 3°C day/night temperature with a 14-h photoperiod.
Biological activity: The activity assay was performed 12 days after sowing for the foliar application of samples at each concentration with a laboratory spray gun. The herbicidal activity of the foliar application was evaluated by visual injury 14 days after treatment (0, no damage; 100, complete control).
Extraction and isolation: A. altissima leaves (5 kg) collected from Nonsan, Korea were dipped in MeOH at room temperature and filtered after three days. After concentrating the methanol, the extract was defatted with hexane, then partitioned between EtOAc and H2O. A portion of the active EtOAc fraction (6 g) was purified by gradient elution on a flash silica gel column using CHCl3:MeOH (50:1, 20:1, 10:1, 5:1, and 100% MeOH). Fraction II [1.03 g, CHCl3-MeOH (20:1)] and fraction III [1.32 g, CHCl3: MeOH (10:1)], which were active in the bioassay, were further purified by ODS Sep-pak cartridge (Alltech, Deerfield, IL, USA) chromatography eluted with an increasing methanol concentration gradient (0%–100%) in water. The active fraction was finally purified by reversed-phase HPLC. Preparative HPLC (C18, 5 μ, 20 × 250 mm; COSMOSIL) was performed using 30%–50% aqueous MeOH, UV detection at 254 nm, a flow rate of 12 mL/min, and gradient elution for 50 min.
Structural analysis: Melting points were measured on a Fisher melting point apparatus and uncorrected. Optical rotations were measured on a Perkin Elmer 341-LC polarimeter. NMR spectra were recorded on a Bruker 900 and 700 spectrometer with standard pulse sequences, operated at 900 and 700 MHz for 1H NMR and 226 and 176 MHz for 13C NMR. Chemical shifts, measured in ppm, were referenced to solvent peaks (δH 2.50 and δC 39.5 for DMSO-d6). HR-TOF-MS spectra were recorded in the positive ESI mode on a Waters Synapt G2 at the Korea Basic Science Institute.
Compound 1 (ailanthone):
1H NMR (DMSO-d6, 900 MHz) δ: 8.23 (1H, s, 11-OH), 7.08 (1H, d, J = 2.8 Hz, 1-OH), 5.99
(1H, d, J = 0.9 Hz, H-3), 5.40 (1H, d, J = 4.5 Hz, 12-OH), 5.05 (1H, d, J = 0.9 Hz, Ha-21), 5.02
(1H, d, J = 0.9 Hz, Hb-21), 4.55 (1H, t, J = 2.8 Hz, H-7), 4.28 (1H, s, H-1), 3.81 (1H, d, J = 9.0
Hz, Ha-20), 3.27 (1H, d, J = 9.0 Hz, Hb-20), 3.67 (1H, d, J = 3.6 Hz, H-12), 2.88 (1H, d, J = 10.8
Hz, H-5), 2.98 (1H, dd, J = 18.0, 14.4 Hz, Ha-15), 2.81 (1H, s, H-9), 2.77 (1H, dd, J = 13.5, 5.4,
H-14), 2.44 (1H, dd, J = 18.0, 5.4 Hz, Hb-15), 2.03 (2H, m, H-6), 1.93 (3H, s, H-18), 1.06 (3H, s,
H-19) ppm; 13C NMR (225 MHz, DMSO-d6): 197.1 (C2), 169.1 (C16), 162.4 (C4), 146.6 (C13),
125.0 (C3), 117.6 (C21), 108.8 (C11), 82.4 (C1), 79.0 (C12), 77.5 (C7), 71.1 (C20), 46.1 (C14),
44.5 (C8 and C10), 43.3 (C9), 41.2 (C5), 34.3 (C15), 25.1 (C6), 22.4 (C18), and 9.5 (C19) ppm;
HRTOFMS (positive ESI mode) m/z 399.1415 [M + Na]+ (calcd for C20H24O7+Na, 399.1420).
Compound 2 (13, 18-dehydroglaucarubinone):
1H NMR (DMSO-d6, 700 MHz) δ: 8.33 (1H, s, 11-OH), 7.16 (1H, d, J = 2.1 Hz, 1-OH), 5.99
(1H, q, J = 0.7 Hz, H-3), 5.64 (1H, br s, H-15), 5.35 (1H, br s, 12-OH), 5.11 (1H, s, 2’-OH), 5.09
(1H, d, J = 1.4 Hz, Ha-21), 4.99 (1H, br s, Hb-21), 4.70 (1H, t, J = 2.8 Hz, H-7), 4.42 (1H, d, J =
2.1 Hz, H-1), 3.81 (1H, d, J = 8.4 Hz, Ha-20), 3.67 (1H, d, J = 4.9 Hz, H-12), 3.32 (1H, dd, J =
8.4 Hz, Hb-20), 3.08 (1H, d, J = 11.9 Hz, H-5), 3.04 (1H, d, J = 11.4 Hz, H-14), 2.97 (1H, s, H-9),
2.06 (2H, m, H-6), 1.93 (3H, s, H-18), 1.70 (1H, dq, J = 140.0, 7.0 Hz, Ha-3’), 1.53 (1H, dq, J =
140.0, 7.0 Hz, Hb-3’), 1.30 (3H, s, H-5’), 1.06 (3H, s, H-19), 0.81 (3H, t, J = 7.0 Hz, H-4’) ppm;
13C NMR (176 MHz, DMSO-d6): 197.0 (C2), 174.2 (C1’), 166.7 (C16), 162.4 (C4), 142.4 (C13),
124.8 (C3), 119.8 (C21), 108.6 (C11), 82.1 (C1), 78.7 (C12), 77.8 (C7), 73.9 (C2’), 70.7 (C20),
68.2 (C15), 50.2 (C14), 46.4 (C8), 44.5 (C10), 44.0 (C9), 40.7 (C5), 32.8 (C3’), 25.8(C5’), 24.6 (C6), 22.2 (C18), 9.5 (C19), and 8.0 (C4’) ppm; HRTOFMS (positive ESI mode) m/z 499.194
[M + Na]+ (calcd for C25H32O9+Na, 499.1944) and m/z 975.3983 [2M + N]+ [calcd for (C25H32O9)2+Na, 975.3990].
Compound 3 (6-α-tigloyloxyailanthone):
1H NMR (DMSO-d6, 700 MHz) δ: 8.15 (1H, s, 11-OH), 7.28 (1H, d, J = 2.8 Hz, 1-OH), 6.91 (dq,
J = 7.7, 2.1 Hz, H3’), 6.02 (1H, s, H-3), 5.55 (1H, d, J = 4.2 Hz, 12- OH), 5.49 (1H, dd, J = 11.9,
2.8 Hz, H-6), 5.02 (1H, d, J = 1.4 Hz, Ha-21), 5.01 (1H, s, Hb-21), 4.63 (1H, d, J = 2.8 Hz, H-7),
4.39 (1H, s, H-1), 3.91 (1H, d, J = 8.4 Hz, Ha-20), 3.69 (1H, t, J = 4.9 Hz, H-12), 3.46 (1H, d, J =
11.9 Hz, H-5), 3.39 (1H, dd, J = 8.4 Hz, Hb-20), 3.01 (1H, dd, J = 18.4, 13.3 Hz, Ha-15), 2.85
(1H, dd, J = 13.3, 5.6 Hz, H-14), 2.83 (1H, s, H-9), 2.47 (1H, dd, J = 18.4, 5.6 Hz, Hb-15), 1.94
(3H, s, H-18), 1.829 (3H, s, H5’), 1.823 (3H, d, J = 4.2 Hz, H4’), 1.21 (3H, s, H-19) ppm; 13C
NMR (176 MHz, DMSO-d6): 196.6 (C2), 168.2 (C16), 165.9 (C1’), 161.2 (C4), 146.1 (C13),
139.2 (C3’), 127.9 (C2’), 127.6 (C3), 117.5 (C21), 109.0 (C11), 82.4 (C1), 79.1 (C12), 77.5 (C7),
70.2 (C20), 67.3 (C6), 47.2 (C10), 46.1 (C14), 45.2 (C8), 44.1 (C5), 42.0 (C9), 34.0 (C15), 24.7 (C18), 14.4 (C4’), 11.9 (C5’), and 10.9 (C19) ppm; HRTOFMS (positive ESI mode) m/z 497.1785 [M + Na]+ (calcd for C25H30O9+Na, 497.1788) and m/z 971.3670 [2M + N]+ [calcd for (C25H30O9)2+Na, 971.3678].
Compound 4 (6-a-tigloyloxychaparrinone):
1H NMR (DMSO-d6, 700 MHz) δ: 8.0 (1H, s, 11-OH), 7.19 (1H, d, J = 2.8 Hz, 1-OH), 6.91 (dq,
J = 7.7, 2.1 Hz, H3’), 6.0 (1H, s, H-3), 5.49 (1H, dd, J = 11.9, 2.8 Hz, H-6), 5.14 (1H, d, J = 4.9
Hz, 12-OH), 4.56 (1H, d, J = 2.8 Hz, H-7), 4.30 (1H, d, J = 2.8 Hz, H-1), 3.93 (1H, d, J = 8.4 Hz,
Ha-20), 3.64 (1H, dd, J = 8.4 Hz, Hb-20), 3.39 (1H, d, J = 11.9 Hz, H-5), 3.21 (1H, t, J = 4.9 Hz,
H-12), 2.72 (1H, dd, J = 18.2, 13.3 Hz, Ha-15), 2.61 (1H, s, H-9), 2.40 (1H, dd, J = 18.2, 5.6 Hz,
Hb-15), 2.09 (1H, m, H-13), 2.05 (1H, m, H-14), 1.94 (3H, s, H-18), 1.82 (3H, br s, H5’), 1.81
(3H, d, J = 7.7 Hz, H4’), 1.22 (3H, s, H-19), 0.85 (3H, d, J = 7.0 Hz, H-21) ppm; 13C NMR (176
MHz, DMSO-d6): 196.5 (C2), 168.9 (C16), 165.8 (C1’), 161.2 (C4), 139.0 (C3’), 127.8 (C2’),
127.5 (C3), 109.1 (C11), 82.5 (C1), 78.1 (C12), 77.6 (C7), 69.3 (C20), 67.3 (C-6), 47.1 (C10),
45.7 (C8), 44.1 (C5), 41.8 (C9), 40.6 (C14), 30.2 (C13), 29.3 (C15), 24.5 (C18), 14.3 (C4’), 12.5 (C21), 11.8 (C5’), and 10.8 (C19) ppm; HRTOFMS (positive ESI mode) m/z 499.1944 [M + Na]+ (calcd for C25H32O9+Na, 499.1944) and m/z 975.3983 [2M + N]+ [calcd for (C25H32O9)2+Na, 975.3990].
Results and Discussion
The MeOH extract of Ailanthus altissima leaves at 10 mg/mL showed 100% control of Sorghum bicolor, Digitaria sanguinalis, Aeschynomene indica, and Abutilon avicennae, and >90% control of Echinochlia crus-galli, Panicum dichotomiflorum, Xanthium strumarium, and Calystegia japonica. Herbicidal activity against Agropyron smithii was slightly weaker (Table 1). The main characteristic was its quick action upon foliar application. External symptoms began to appear within 24 h of exposure, and the weeds were completely controlled five days after treatment. MeOH extract showing herbicidal activity was purified by ethyl acetate fractionation and a series of chromatographic techniques, including silica gel column, C18 Sep- pak cartridge, Sephadex LH20, and preparative High-Performance Liquid Chromatography (HPLC). Four quassinoids, 1-4 (Figure 1), were isolated from the MeOH extract of Ailanthus altissima leaves and showed herbicidal activity of 98, 95, 85, and 75%, respectively, when applied to D. sanguiinalis at a concentration of 10 μg/mL (Table 2 and Figure 2).
| Rates (mg/mL) | Herbicidal activity (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SORBI | ECHCG | AGRSM | DIGSA | PANDI | SOLNI | AESIN | ABUTH | XANSI | CAGHE | |
| 10 | 100 | 95 | 30 | 100 | 98 | 70 | 100 | 100 | 90 | 95 |
| 5 | 40 | 40 | 20 | 100 | 70 | 60 | 100 | 80 | 40 | 70 |
| 2.5 | 40 | 30 | 10 | 90 | 20 | 20 | 98 | 98 | 40 | 70 |
| 1 | 40 | 30 | 10 | 70 | 20 | 20 | 95 | 98 | 40 | 70 |
Note: SORBI: Sorghum bicolor, ECHCG: Echinochloa crus-galli, AGRSM: Agropyron smithii, DIGSA: Digitaria sanguinalis, PANDI: Panicum dichotomiflorum, SOLNI: Solanum nigrum, AESIN: Aeschynomene indica, ABUTH: Abutilon avicennae, XANSI: Xanthium strumarium, CAGHE: Calystegia japonica.
Table 1: Herbicidal activity of foliar application of methanol extracts from Ailanthus altissima to several weeds in a greenhouse condition.
| Herbicidal activity at 10 μg/mL (%) | |
|---|---|
| Compound 1 | 98 |
| Compound 2 | 95 |
| Compound 3 | 85 |
| Compound 4 | 75 |
Table 2: Herbicidal activity of isolated compounds against Digitaria sanguinalis.
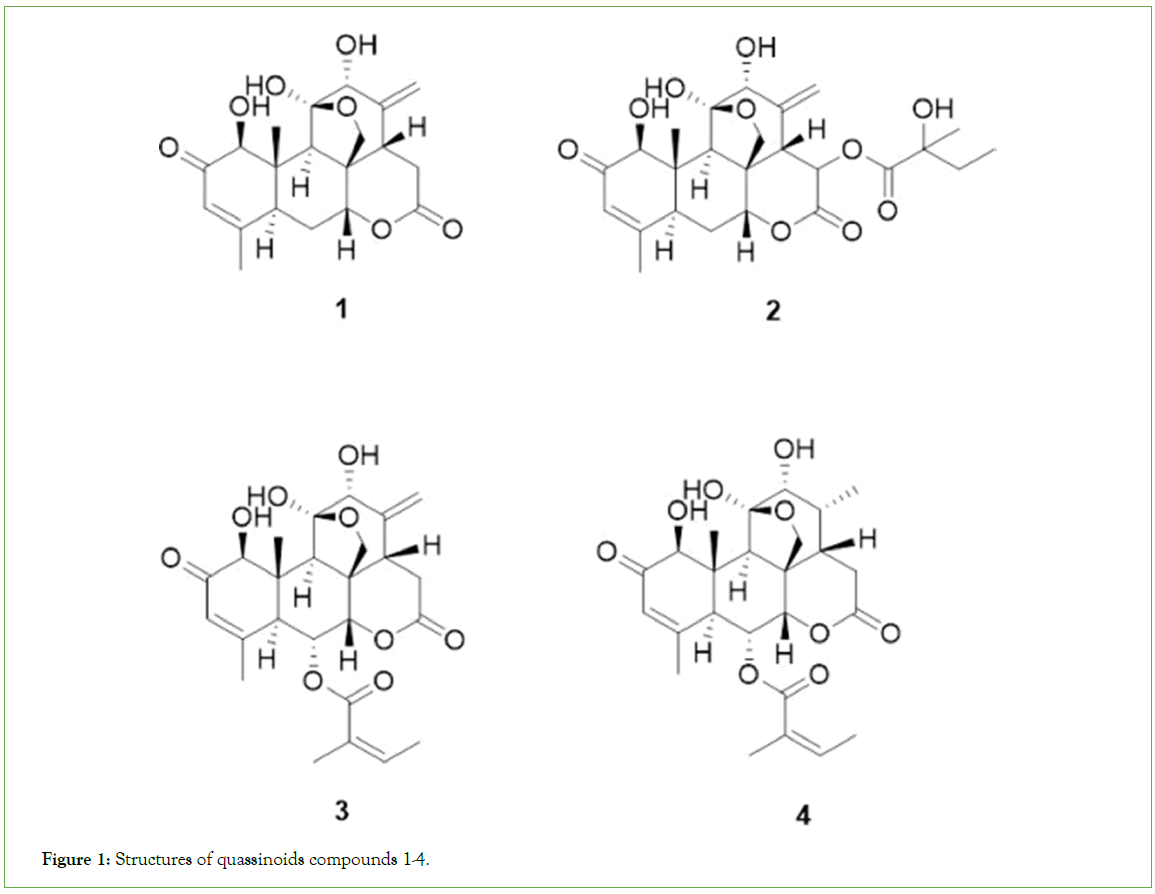
Figure 1: Structures of quassinoids compounds 1-4.
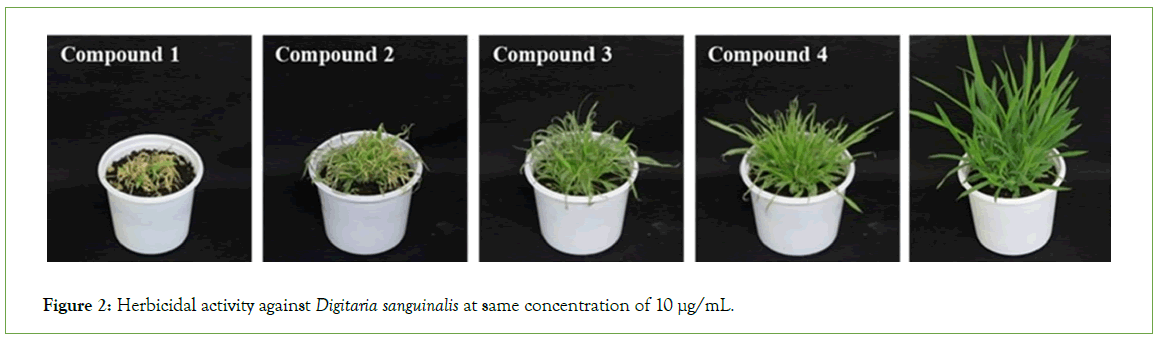
Figure 2: Herbicidal activity against Digitaria sanguinalis at same concentration of 10 μg/mL.
Among these, 3 was a new compound, and the characterization and spectroscopic analysis of this compound were performed by data comparison with literature values [14]. The 1H and 13C NMR (nuclear magnetic resonance) data for these compounds are shown in the experimental section. The 1H (Table 3) and 13C NMR (Table 4) signals assignments were aided by HSQC (Heteronuclear Single Quantum Correlation), HMBC (Heteronuclear Multiple Bond Correlation), and 1H-1H COSY (Correlation Spectroscop Y) experiments. The other compounds were identified as ailanthone (1) [15], 13, 18-dehydroglaucarubinone (2) [16], and 6-α-tigloyloxychaparrinone (4) [17,18]. Compound 3 was obtained as a white solid. The molecular formula was established as C25H32O9 based on a quasi-molecular ion at m/z 497.1785 [M + Na]+ (calcd 497.1788) in its HRTOFMS. The 1H NMR spectrum of 3 displayed signals for two olefinic protons [δH 6.91 (1H, dq, J = 7.7, 2.1, H-3’), 6.02 (1H, s, H-3)], an exo-methylene [δH 5.02 (1H, d, J = 2.1, Ha-21), 5.01 (1H, s, Hb-21)], four oxygenated methines [δH 5.49 (1H, dd, J = 11.9, 2.8, H-6), 4.63 (1H, d, J = 2.8, H-7), 4.39 (1H, s, H-1), 3.69 (1H, d, J = 3.5, H-12)], one oxygenated methylene [δH 3.91 and 3.39 (each 1H, d, J = 8.4, H-20)], one methylene proton [δH 3.01 (1H, dd, J = 18.4, 13.3, Ha-15) and 2.40 (1H, dd, J = 18.4, 5.6, Hb-15)] and four methyl groups [δH 1.94 (3H, s, H-18), 1.829 (3H, br s, H-5’), 1.823 (3H, d, J = 7.7, H-4’), and 1.21 (3H, s, H-19)]. The 13C NMR spectrum exhibited 25 carbon signals, including three carbonyl signals, six olefinic carbon signals, one hemiketal carbon signal, four methyl carbon signals, two methylene carbon signals, seven methine carbon signals, and two quaternary carbon signals. Comparison of the NMR data of 3 with those of ailanthone (1) revealed that the two compounds possessed the same quassinoid moiety, except that the C6 position in ailanthone (1) was substituted with a tigloyloxy group in 3. The 1H-1H COSY between H-3’ (δH 6.91) and H-4’ (δH 1.823) and the HMBC correlation between H-3’ (δH 6.91), H-5’ (δH 1.829), and C1’ (δC 165.8) and H-4’ (δH 1.81) and C-2’ (δC 127.8) confirmed the existence of a tigloyloxy group and that it was connected to the C6 position of 3, established by a long-range correlation between δH 5.49 (H-6 of quassinoid moiety) and carboxylic carbon (δC 165.9, C1’ of tigloyloxy group) in the HMBC spectrum of 3. The correlations between H-9 and H-1/H-5 and between H-7 and H-14 in the ROESY spectrum suggested that the configuration of 3 was the same as that of ailanthone (1). And the ROESY correlation between H-3’ and H-5’ indicated that the geometry of the double-bond of the tigloyloxy group was Z-form. Thus, the structure of compound 3 was determined to be 6-α- tigloyloxyailanthone. The relative stereochemistry of 3 was confirmed from its 2D-ROESY NMR spectrum. The NOE correlations are shown by arrows in Figure 3 (2D-ROESY).
| Proton | Compound | |||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| H-1 | 4.28(s) | 4.42 (d, 2.1) | 4.39 (s) | 4.30 (d, 2.8) |
| H-3 | 5.99 (d, 0.9) | 5.99 (q, 0.7) | 6.02 (s) | 6.0 (s) |
| H-5 | 2.88 (d, 10.8) | 3.08 (d, 11.4) | 3.46 (d, 11.9) | 3.39 (d, 11.9) |
| H-6 | 2.03 (m) | 2.06 (m) | 5.49 (dd, 11.9,2.8) | 5.49 (dd, 11.9, 2.8) |
| H-7 | 4.55 (t, 2.8) | 4.70 (t, 2.8) | 4.63 (d, 2.8) | 4.56 (d, 2.8) |
| H-9 | 2.81 (s) | 2.97(s) | 2.83(s) | 2.61(s) |
| H-12 | 3.67 (d, 3.6) | 3.67 (d, 4.9) | 3.69 (d, 3.5) | 3.21 (t, 4.9) |
| H-13 | - | - | - | 20.9 (m) |
| H-14 | 2.77 (dd, 13.5, 5.4) | 3.04 (d, 11.4) | 2.85 (dd, 13.3, 5.6) | 2.05 (m) |
| Ha-15 | 2.98 (dd, 18.4, 13.5) | 5.64 (br s) | 3.01 (dd, 18.4, 13.3) | 2.72 (dd, 18.2, 13.3) |
| Hb-15 | 2.44 (dd, 18.4, 5.4) | 2.47 (dd, 18.4, 5.6 ) | 2.40 (dd, 18.2, 5.6) | |
| H-18 | 1.93 (s) | 1.93 (s) | 1.94 (s) | 1.94 (s) |
| H-19 | 1.06 (s) | 1.06 (s) | 1.21 (s) | 1.22 (s) |
| Ha-20 | 3.81 (d, 9.0) | 3.81 (d, 8.4) | 3.91 (d, 8.4) | 3.93 (d, 8.4) |
| Hb-20 | 3.27 (d, 9.0) | 3.32 (d, 8.4) | 3.39 (d, 8.4) | 3.64 (d, 8.4) |
| Ha-21 | 5.05 (d, 0.9) | 5.09 (d, 1.4) | 5.02 (d, 1.4) | - |
| Hb-21 | 5.02 (d, 0.9) | 4.99 (br s) | 5.01 (br s) | - |
| Ha-3' | - | 1.70 (dq, 14.0,7.0) | 6.91 (dq, 7.7,2.1) | 6.91 (dq, 7.7, 2.1) |
| Hb-3' | - | 1.53 (dq, 14.0,7.0) | - | - |
| H-4' | - | 0.81 (t, 7.0) | 1.823 (d, 4.2) | 1.81 (d, 7.7) |
| H-5' | - | 1.30 (s) | 1.829 (s) | 1.82 (br s) |
| 1-OH | 7.08 (s) | 7.16 (d, 2.1) | 7.28 (s) | 7.19 (d, 2.8) |
| 11-OH | 8.23 (s) | 8.33 (s) | 8.15 (s) | 8.0 (s) |
| 12-OH | 5.40 (d, 4.5) | 5.35 (br s) | 5.55 (d, 4.2) | 5.14 (d, 4.9) |
Table 3: 1H-NMR Data (DMSO-d6) for compounds 1-4.
| Carbon | Compound | |||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| C-1 | 82.4 | 82.1 | 82.4 | 82.5 |
| C-2 | 197.1 | 197 | 196.6 | 196.5 |
| C-3 | 125 | 124.8 | 127.6 | 127.5 |
| C-4 | 162.4 | 162.4 | 161.1 | 161.2 |
| C-5 | 41.2 | 40.7 | 44.1 | 44.1 |
| C-6 | 25.1 | 24.6 | 67.3 | 67.3 |
| C-7 | 77.5 | 77.8 | 77.6 | 77.6 |
| C-8 | 44.5 | 46.4 | 45.2 | 45.7 |
| C-9 | 43.3 | 44 | 42 | 41.8 |
| C-10 | 44.5 | 44.5 | 47.2 | 47.1 |
| C-11 | 108.8 | 108.6 | 109 | 109.1 |
| C-12 | 79 | 78.7 | 79.1 | 78.1 |
| C-13 | 146.6 | 142.4 | 146.1 | 30.2 |
| C-14 | 46.1 | 50.2 | 46.1 | 40.6 |
| C-15 | 34.3 | 68.2 | 34 | 29.3 |
| C-16 | 169.1 | 166.7 | 168.2 | 168.9 |
| C-17 | 22.4 | 22.2 | 24.7 | 24.5 |
| C-18 | 9.5 | 9.5 | 10.9 | 10.8 |
| C-19 | 71.1 | 70.7 | 70.2 | 69.3 |
| C-20 | 117.6 | 119.8 | 117.5 | 12.5 |
| C-1' | - | 174.2 | 165.9 | 165.8 |
| C-2' | - | 73.9 | 127.9 | 127.8 |
| C-3' | - | 32.8 | 139.2 | 139 |
| C-4' | - | 8 | 14.4 | 14.3 |
| C-5' | - | 25.8 | 11.9 | 11.8 |
Table 4: 13C-NMR Data (DMSO-d6) for compounds 1-4.
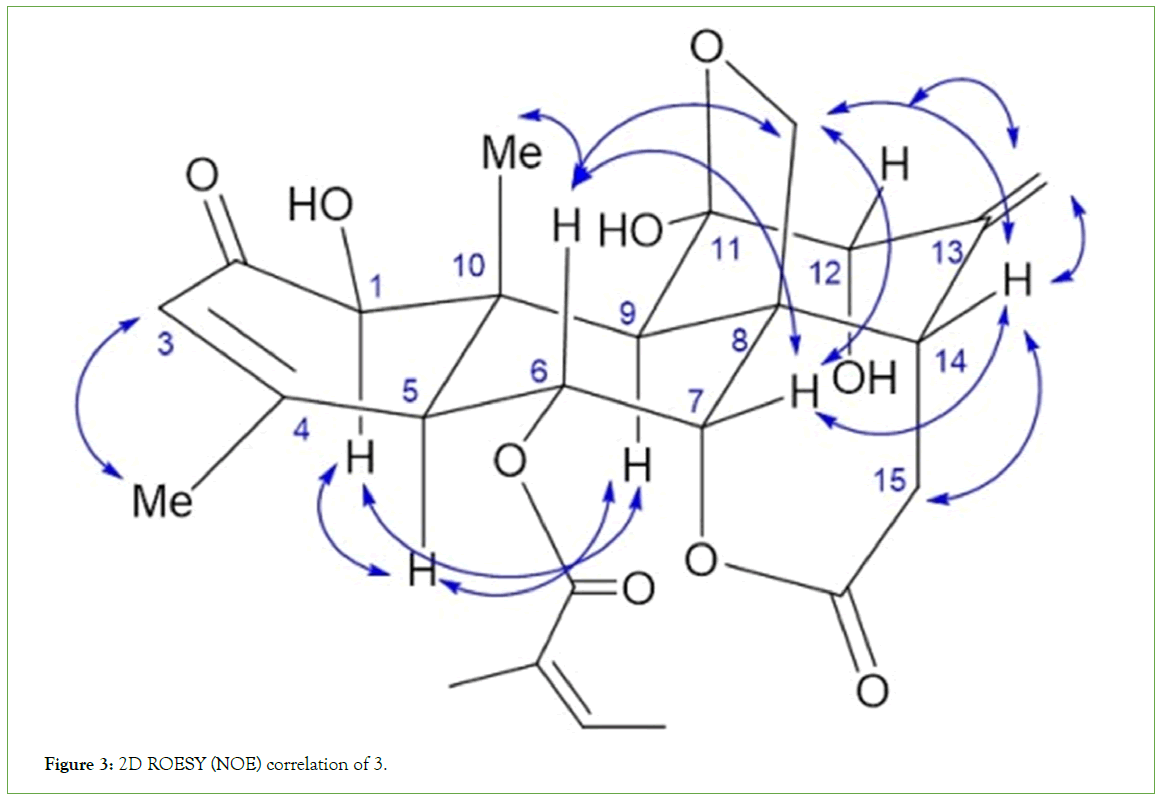
Figure 3: 2D ROESY (NOE) correlation of 3.
Conclusion
Herbicidal activity of methanol extract of A. altissima leaves with foliar application on D. sanguiinalis at 2, 5, and 10 mg mL-1 was 90, 100 and 100%, respectively, the main herbicidal symptoms were chlorosis or burn-down and followed by necrosis and eventual death. On 10 weed species, the methanol extract showed strong herbicidal activity and it showed excellent herbicidal activity particularly on A. indica and A. avicennae even at the lowest concentration of 1 mg mL-1. Three known quassinoid compounds (1, 2, 4) and one unknown compound (3) isolated from methanol extract showed herbicidal activity of 98, 95, 75, and 85%, respectively, against D. sanguinalis at 10 μ mL-1. Compound 3, a white solid, was identified as 6-α- tigloyloxyailanthone with a molecular formula of C25H32O9 by the analyses of HR-TOF-MS and 1H and 13C NMR and 2D NMR spectral data.
This study showed that A. altissima extracts have potential as bioherbicide and one unknown compounds may be used as lead compounds for development of new herbicides.
Supporting Information
Structures and spectra of compound 1, 2, 3 and 4 (Figures S1-S16).
Author Contributions
The authors confirm contribution to the paper as follows: study conception and design: Y. S. Kim, S. S. Moon; data collection: J. S. Choi, S. S. Moon; analysis and interpretation of results: Y. S. Kim, S. S. Moon, J. S. Choi; draft manuscript preparation: Y. S. Kim, J. S. Choi. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
This work was carried out with the support of the Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ0161282023), Rural Development Administration, Republic of Korea.
Competing Interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Cormier MJ, Flasch H, Franck B, Hori K, Jaenicke L, Keller-Schierlein W, et al. Quassinoid bitter principles. Fortschr Chem Org Naturs.1973:101-150.
[Crossref] [Google Scholar] [PubMed]
- Vieira IJ, Braz-Filho R. Quassinoids: structural diversity, biological activity and synthetic studies. Stud Nat Prod. 2006;33:433-492.
- Guo Z, Vangapandu S, Sindelar RW, Walker LA, Sindelar RD. Biologically active quassinoids and their chemistry: Potential leads for drug design. Curr Med chem. 2005;12(2):173-190.
[Crossref] [Google Scholar] [PubMed]
- Alves IA, Miranda HM, Soares LA, Randau KP. Simaroubaceae family: Botany, chemical composition and biological activities. Rev Bras Farmacogn 2014;24:481-501.
- Elson S, Howe I, Jarman M, McDougal PG, Polonsky J, Schmuff NR, et al. Quassinoid bitter principles II. Fortschr Chem Org Naturst.1985:221-264.
[Crossref] [Google Scholar] [PubMed]
- Fukamiya N, Okano M, Miyamoto M, Tagahara K, Lee KH. Antitumor agents, 127. Bruceoside C, A new cytotoxic quassinoid glucoside, and related compounds from Brucea javanica. J Nat Prod. 1992;55(4):468-475.
[Crossref] [Google Scholar] [PubMed]
- Ang HH, Chan KL, Mak JW. In vitro antimalarial activity of quassinoids from Eurycoma longifolia against Malaysian chloroquine-resistant Plasmodium falciparum isolates. Planta Medica. 1995;61(02):177-178.
[Crossref] [Google Scholar] [PubMed]
- Apers S, Cimanga K, Berghe DV, Van Meenen E, Longanga AO, Foriers A, et al. Antiviral activity of simalikalactone D, a quassinoid from Quassia africana. Planta Med. 2002;68(01):20-24.
[Crossref] [Google Scholar] [PubMed]
- Silva RL, Lopes AH, França RO, Vieira SM, Silva EC, Amorim RC, et al. The quassinoid isobrucein B reduces inflammatory hyperalgesia and cytokine production by post-transcriptional modulation. J Nat Prod. 2015;78(2):241-249.
[Crossref] [Google Scholar] [PubMed]
- Leskinen V, Polonsky J, Bhatnagar S. Antifeedant activity of quassinoids. J Chem Ecol. 1984;10(10):1497-1507.
[Crossref] [Google Scholar] [PubMed]
- Daido M, Fukamiya N, Okano M, Tagahara K, Hatakoshi M, Yamazaki H, et al. Antifeedant and insecticidal activity of quassinoids against diamondback moth (Plutella xylostella). Biosci Biotechnol Biochem. 1993;57(2):244-246.
[Crossref] [Google Scholar] [PubMed]
- Tada H, Yasuda F, Otani K, Doteuchi M, Ishihara Y, Shiro M, et al. New antiulcer quassinoids from Eurycoma longifolia. Eur J Med Chem 1991;26:345-349.
- Heisey RM, Kish Heisey T. Herbicidal effects under field conditions of Ailanthus altissima bark extract, which contains ailanthone. Plant Soil. 2003;256:85-99.
- Kubota K, Fukamiya N, Tokuda H, Nishino H, Tagahara K, Lee KH et al. Quassinoids as inhibitors of Epstein-Barr virus early antigen activation. Cancer Letters 1997;113()1-2:165-168.
[Crossref] [Google Scholar] [PubMed]
- Kubota K, Fukamiya N, Hamada T, Okano M, Tagahara K, Lee KH, et al. Two new quassinoids, ailantinols A and B, and related compounds from Ailanthus altissima. J Nat Prod 1996;59(7):683-686.
[Crossref] [Google Scholar] [PubMed]
- Polonsky J, Varon Z, Jacquemin H, Pettit GR. The isolation and structure of 13, 18-dehydroglaucarubinone, a new antineoplastic quassinoid from Simarouba amara. Experientia. 1978;34:1122-1123.
[Crossref] [Google Scholar] [PubMed]
- Carter CA, Tinto WF, Reynolds WF, McLean S. Quassinoids from Quassia multiflora: Structural assignments by 2D Nmr spectroscopy. J Nat Prod.1993;56(1):130-133.
- Okunade AL, Bikoff RE, Casper SJ, Oksman A, Goldberg DE, Lewis WH, et al. Antiplasmodial activity of extracts and quassinoids isolated from seedlings of Ailanthus altissima (Simaroubaceae). Phytotherapy Research 2003;17(6):675-677.
[Crossref] [Google Scholar] [PubMed]
Citation: Kim YS, Moon SS, Choi JS (2023) Herbicidal Quassinoids Isolated from Ailanthus altissima Leaves. Biochem Anal Biochem. 12:517.
Copyright: © 2023 Kim YS, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.


