Indexed In
- Open J Gate
- Genamics JournalSeek
- ResearchBible
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- Publons
- MIAR
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Mini Review - (2022) Volume 11, Issue 2
COVID-19: Review on the Biochemical Perspective of the Structural and Non-structural Proteins Involved in SARS-CoV-2 Corona Virus
Soumya V. Menon*Received: 10-Feb-2022, Manuscript No. BABCR-22-15624; Editor assigned: 14-Feb-2022, Pre QC No. BABCR-22-15624 (PQ); Reviewed: 25-Feb-2022, QC No. BABCR-22-15624; Revised: 02-Mar-2022, Manuscript No. BABCR-22-15624 (R); Published: 07-Mar-2022, DOI: 10.35248/2161-1009.22.11.420
Abstract
A novel member of human SARS-CoV-2 that has recently emerged in Wuhan, China, is now formally named as SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2). This is a unique strain of RNA viruses that have not been previously observed in humans. The virus has wide host adaptability and is capable of causing severe diseases in humans, masked palm civets, mice, dogs, cats, camels, pigs, chickens, and bats. The SARS-CoV-2 typically causes respiratory and gastrointestinal sickness in both humans and animals. The main objective of this review is to address the important biochemical aspects of SARS-CoV-2 corona virus. There is a critical need to study the structural and non-structural proteins involved in this virus to understand and support the clinical, biochemical and structural studies to combat the pandemic occurrence all over the world by SARS-CoV-2. The review also gives a comprehensive view of the various analytical techniques used to study the corona virus such as size exclusion chromatography, circular dichroic spectroscopy, and Multiage Light Scattering and micro array methods.
Keywords
Circular dichroism; Corona virus; Respiratory syndrome; SARS-CoV-2
Introduction
Coronaviruses (CoVs) are the largest group of viruses belonging to the Nidovirales order, which includes Coronaviridae, Arteriviridae, Mesoniviridae, and Roniviridae families. The Coronaviridae are further subdivided into four genera, the alpha, beta, gamma, and delta coronaviruses. The viruses were initially sorted into these genera based on serology but are now divided by phylogenetic clustering. All viruses belonging to Nidovirales order are enveloped, nonsegmented positive-sense RNA viruses. They all contain very large genomes for RNA viruses, with some viruses having the largest identified RNA genomes, containing up to 33.5 kilo base (kb) genomes. Other common features within the Nidovirales order include: (1) a highly conserved genomic organization, with a large replicase gene preceding structural and accessory genes.
When we see the history of corona virus, or SARS CoV, the epidemic was out broken once in 2002 and it was controlled in 2003 and the virus has not since returned but, a novel human CoV emerged in the Middle East in 2012. This virus, named Middle East Respiratory Syndrome-CoV (MERS-CoV), was found to be the causative agent in a series of highly pathogenic respiratory tract infections in Saudi Arabia and other countries in the Middle East [1]. Based on the high mortality rate of ~50% in the early stages of the outbreak, it was feared the virus would lead to a very serious outbreak. However, the outbreak did not accelerate in 2013, although sporadic cases continued throughout the rest of the year. In April 2014, a spike of over 200 cases and almost 40 deaths occurred, prompting fears that the virus had mutated and was more capable of human-to human transmission. More likely, the increased number of cases resulted from improved detection and reporting methods combined with a seasonal increase in birthing camels. As of August 27th, 2014 there have been a total of 855 cases of MERS-CoV, with 333 deaths and a case fatality rate of nearly 40%, according to the European Center for Disease Prevention and Control. Later followed by the month of December 2019, a novel infectious one that was named as COVID-19 was identified in the Wuhan city of Hubei province, China.
On the initial monitoring of some samples of a cluster of patients admitted with fever, cough and shortness of breath revealed pneumonia of unknown origin, later proved to be a viral infection by pathogen belonging to Betacoronavirus B lineage. Based on genetic similarities to the genome of Middle East Respiratory Syndrome Virus (MERS-CoV), Severe Acute Respiratory Syndrome Virus (SARS-CoV) and bat coronavirus RaTG13, this novel virus was named SARS-CoV-2 [2]. Bats that have been considered the natural habitat for various coronaviruses including SARS-CoV- like and MERS-CoV-like viruses were predicted, based on the results of genome sequencing and evolutionary study, to be the suspected natural host in the origin of the SARS-CoV-2 [3,4]. There are different types of coronavirus that cause infections in human beings and SARS-CoV-2 is seventh in this series. There are four different genera of coronavirus: α-CoV, βCoV, γ-CoV, and δ-CoV. The α and β-CoV cause infection in mammals, whereas γ and δ-CoV infect the birds. SARS-CoV-2 on the other hand is more potent than SARS-CoV and MERS-CoV, and can lead to death due to pneumonia [5]. COVID-19 was shown exhibiting high viral shedding in the upper respiratory tract at an early stage of infection, resulting in a high proportion of transmission-competent individuals that are pre-symptomatic, asymptomatic, and mildly symptomatic. In this review, an overview is given on the various aspects and views to SARS CoV-2, mainly the various structural and nonstructural proteins, different analytical techniques involved in research studies related to SARS-CoV-2 which gives an outline about the pandemic (Figure 1).
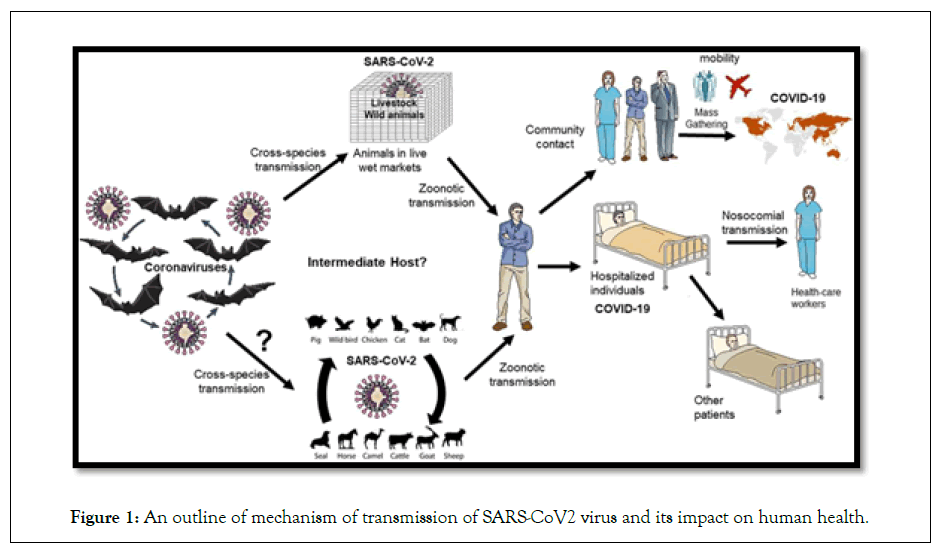
Figure 1: An outline of mechanism of transmission of SARS-CoV2 virus and its impact on human health.
Biochemical Characterization of Sars-Cov-2 Virus
The infection caused by SARS-CoV-2 could use a one kind of host protein named Angiotensin-Converting Enzyme 2 (ACE2) as used by the SARS-CoV, to infect humans [6]. ACE2 is expressed by many tissues, such as the epithelial cells lining the nose, mouth, and lungs and acts as a major entry point for the COVID-19 virus. Latest studies by Hoffman, indicated that SARS-CoV-2 utilizes a cell surface complex that comprises a primary receptor called ACE2 and a serine protease named Trans Membrane Protease Serine 2 (TMPRSS2) co-localized on host cell surfaces to enhance the spike (S) protein-mediated entry of SARS-CoV-2 to the host cell (Figure 2) [7].
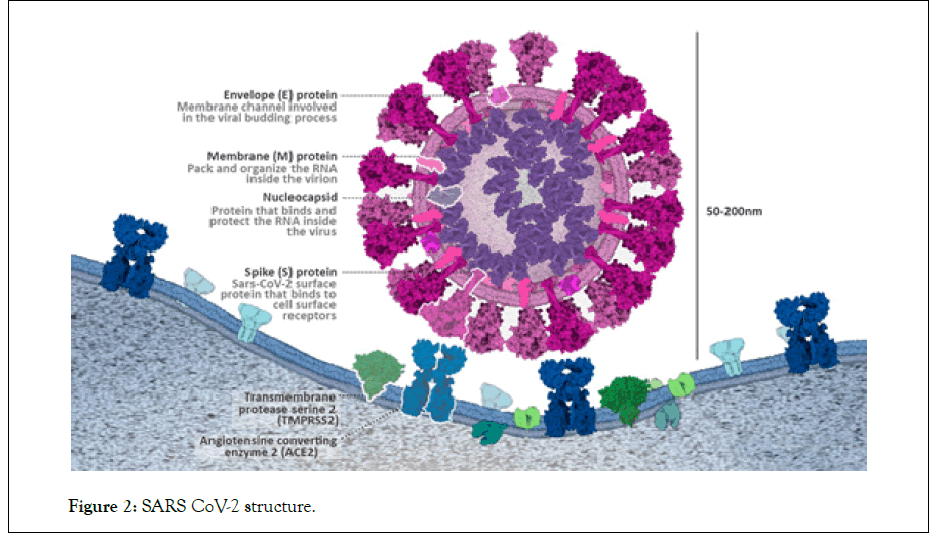
Figure 2: SARS CoV-2 structure.
The SARS-CoV-2 genome is composed of approximately about 30,000 nucleotides, which encodes four structural proteins which includes nucleocapsid (N) protein, envelope (E) protein, spike (S) protein, and envelope (E) protein and membrane (M) protein [4]. Among all of these proteins, N protein is abundantly expressed and is highly immunogenic for the period of infection [3,8]. Moreover, N protein is regularly applied in the development of vaccines and in serological assays. Based on the results from serological assay, it was concluded by Ahirwar et al. That the specific antibodies against the N protein in the serum of SARS patients have higher sensitivity and longer persistence than those of other structural proteins of SARS-CoV-2. Moreover, anti-N antibodies have been detected with high specificity in the early stage of infection. Very less and meager studies are still available for this SARS-CoV-2 N protein. Thus, any information generated from the analysis of this protein, both in vivo and in vitro, will improve our understanding of COVID-19 and help us to design better biological agents for the treatment or diagnostics of this pandemic so that future screening and diagnosis will be easier.
The domain structure of SARS CoV2 N protein was found in the study by Zeng [9,10]. In a similar line of research, some scientists all over the world have successfully performed the expression and purification of two recently reported recombinant versions of the S protein (OptSpike1 and OptSpike2) and this construct has additionally been utilized as an antigen for clinical ELISAs [11,12].
The chemistry of the various nonstructural proteins (NSP3, NSP5, NSP12, NSP13, NSP14, NSP15, NSP16) of SARS CoV-2 was discussed by Yoshimoto [13]. Secondly, a recent major focus of this pandemic is the variant strains of SARS CoV-2 that are increasingly occurring and more transmissible. For example, “D614G”, strain possessed a glycine (G) instead of an aspartate (D) at position 614 of the spike protein. Additionally, other emerging strains called “501Y.V1” and “501Y.V2” have several differences in the receptor binding domain of the spike protein (N501Y) as well as other locations. These structural changes may enhance the interaction between the spike protein and the ACE2 receptor of the host, increasing infectivity according to Yoshimoto.
Some of the nonstructural proteins called enzymes such as NSP13, guanosine N7-methyltransferase: NSP14, an endoribonuclease: NSP15, and a 2′-O-ribose-methyltransferase (NSP16) are also identified recently. This group of enzymes can generally be classified into different subgroups: (i) proteases (NSP3 and NSP5), (ii) enzymes involved in the 5′-capping modification of viral RNA—a posttranslational modification of viral RNA (to allow the viral RNA to escape the host innate immune system) (NSP13, NSP14, NSP15, and NSP16) (Figure 3).
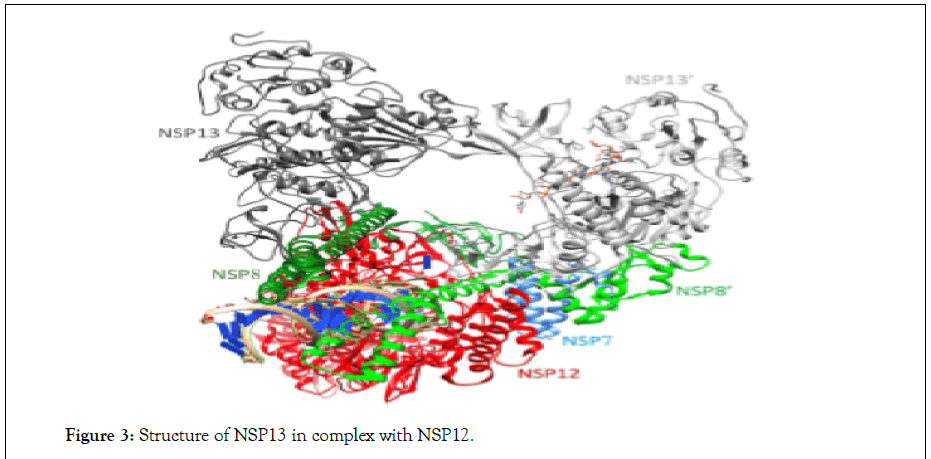
Figure 3: Structure of NSP13 in complex with NSP12.
Analytical Techniques Used the Diagnosis of Sars Cov-2
Accurate, affordable, and rapid testing holds key importance in timely diagnosis and isolation of infected individuals to curtail SARS-CoV-2 spread from both symptomatic and asymptomatic viral carriers. Different types of COVID-19 tests, including mainly the Reverse-Transcription Polymerase Chain Reaction (RT-PCR) for detection of viral RNA, lateral flow immuno chromatographic strips for automated rapid antigen test, chest CT scan for analysis of associated clinical symptoms, and antibody test for analysis of SARS-CoV-2 antibodies have been used to detect the presence of SARS-CoV-2 or the body’s response to the infection. Besides these, methods based on the use of electrochemical detection systems, isothermal nucleic acid amplification, nucleic acid microarrays, high-throughput sequencing, and serological and immunological assays based on colorimetry, fluorescence, and luminescence are also being developed to improve sensitivity, affordability, and diagnostic capacity for mass testing and assist in preventing future epidemics [9,13].
In order to characterize the conformational properties of the N protein, Zeng et al [9]. employed Circular Dichroism (CD) spectroscopy to analyze the secondary structures. The technique used for expression and characterization of S protein of SARS CoV2, were Size Exclusion Chromatography and Multiangle Light Scattering (SEC-MALS).
Conclusion
More biochemical techniques need to be explored in studying the different aspects of SARS-CoV-2 virus. This may enhance the rapid testing methods, diagnosis and even prevention of occurrence of new and menacing strains of SARS-CoV-2 corona virus.
REFERENCES
- Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Corona. 2015:1-23.
[Crossref Med], [Google Scholar], [PubMed].
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nat. 2020;579(798):270-273. [Crossref Med],
[Google Scholar], [PubMed].
- Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, et al. Bats are natural reservoirs of SARS-like coronaviruses. Sci. 2005;310(5748):676-679.
[Crossref Med], [Google Scholar], [PubMed].
- Banerjee A, Kulcsar K, Misra V, Frieman M, Mossman K. Bats and coronaviruses. Viruses. 2019;11(1):41.
[Crossref Med], [Google Scholar], [PubMed].
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Sci. 2020;367(6483):1260-1263. [Crossref Med],
[Google Scholar], [PubMed].
- Hoffmann M, Kleine-Weber H, Krüger N, Müller M, Drosten C, Pöhlmann S. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. BioRxiv. 2020. [Crossref Med],
[Google Scholar], [PubMed].
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020. [Crossref Med],
[Google Scholar], [PubMed].
- Ahirwar R, Gandhi S, Komal K, Dhaniya G, Tripathi PP, Shingatgeri VM, et al. Biochemical composition, transmission and diagnosis of SARS-CoV-2. Biosci Rep. 2021;41(8). [Crossref Med],
[Google Scholar], [PubMed].
- Zeng W, Liu G, Ma H, Zhao D, Yang Y, Liu M, et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem Biophys Res Commun. 2020;527(3):618-623. [Crossref Med],
[Google Scholar], [PubMed].
- Amanat F, Stadlbauer D, Strohmeier S, Nguyen TH, Chromikova V, McMahon M, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020;26(7):1033-1036. [Crossref Med],
[Google Scholar], [PubMed].
- Bortz III RH, Florez C, Laudermilch E, Wirchnianski AS, Lasso G, Malonis RJ, et al. Development, clinical translation, and utility of a COVID-19 antibody test with qualitative and quantitative readouts. medRxiv. 2020. [Crossref Med],
[Google Scholar], [PubMed].
- Herrera NG, Morano NC, Celikgil A, Georgiev GI, Malonis RJ, Lee JH, et al. Characterization of the SARS-CoV-2 S protein: Biophysical, biochemical, structural, and antigenic analysis. ACS Omega. 6:85-102. [Crossref Med],
[Google Scholar], [PubMed].
- Yoshimoto FK. A Biochemical Perspective of the Nonstructural Proteins (NSPs) and the Spike Protein of SARS CoV-2. Proteins. 2021;40(3):260-95. [Crossref Med],
[Google Scholar], [PubMed].
Citation: Menon SV (2022) COVID-19: Review on the Biochemical Perspective of the Structural and Non-structural Proteins Involved in SARS- CoV-2 Corona Virus. Biochem Anal Biochem. 11:420.
Copyright: © 2022 Menon SV. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

