Indexed In
- Open J Gate
- Genamics JournalSeek
- ResearchBible
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- Publons
- MIAR
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Review Article - (2023) Volume 12, Issue 3
A Review on Molecular Dynamics Simulation in Nano and Micro-sized Biomolecular Structures in Biological Aspects: Its Advancement and Advances
Sanjay Kannan*Received: 23-Jun-2023, Manuscript No. BABCR-23-21929; Editor assigned: 27-Jun-2023, Pre QC No. BABCR-23-21929 (PQ); Reviewed: 12-Jul-2023, QC No. BABCR-23-21929; Revised: 20-Jul-2023, Manuscript No. BABCR-23-21929 (R); Published: 28-Jul-2023, DOI: 10.35248/2161-1009.23.12.492
Abstract
Molecular dynamics is an essential role over the biological aspects in the technology of science. Biological experiments deal with the identifying and analyzing the biomolecular structures includes movement of the individual molecules over the period of time. Biological aspects include proteins, DNA, bioinformatics, drugs, chemicals, medicines and even in the food processing. Biomolecular structures include cells of nano and micro sized cells and even in angstrom. Though the going with the experiment is slight easier when compared to theory and simulations, there are some important limitations includes, it difficult to observe the individual cells though it is possible it is at low resolution and also makes difficult to calculate the forces and motions in theory. So, simulation makes the work easier and faster, doesn’t matter the structure of biomolecules is simple or complex and able to provide the exact result of the individual molecules or cell from refine high resolution. Molecular dynamics make itself a required one in every sectors mainly for researchers which makes them to observe the even a nano-sized atoms/molecules at refined resolutions. Molecular dynamics is the technology of dealing and simulating the motions of the system of molecules and applied to biomolecules which gives the variation in the comparative positions of the atoms or molecules in DNA or proteins over a time.
Keywords
Molecular dynamics; Biological aspects; Biomolecular structures; Proteins; DNA’s; Ligands; Drug discovery
Introduction
Molecular dynamics is the advanced technology of where the motion of the particles can be simulated. The motion of the particle can be applicable to systems as minute as atoms, even as congregation and the molecules that undertakes chemical reactions [1]. In every case, the particles that were being simulated through molecular dynamics simulation require strong impression of the contact for the particles by which the forces and equations of the motion which governs the movement of particles can be calculated [2]. But the interaction between the atoms and molecules will differ from simulating of simple particles to the complex particles. Also, simulation will provide the detailed information of every individual particle over the period of time. So, the Molecular Dynamics simulation can be used to understand the characteristics of the system of models which is more simple and easier when compared to the actual system of experiments. Although the results from the experiments have vital role in the methodology, contrast between the experiment and simulation provides an exact result and also will provide principles for improving the methodology [3]. This is predominantly significant because theoretical estimations of systematic errors intrinsic in simulations is impossible i.e., the errors that are found on the experimental capacities are difficult to enumerate. Two qualities of molecular dynamics simulation had a vigorous role in the unpredictable development and wide range of applications [4]. Simulation will provide the 0020 complete data includes motion, alignment etc. of every atom which is easy rather going with experiments. Another advantage in that, though the parameters used in simulation are appropriate they are totally under the control of user so that adding or removing specific parameters altering the exact contributions their character in given property can be analysed [5].
Literature Review
Molecular dynamics is not simple antique spectacle, but it is quickly emerging field of science especially in the field of biological aspects that includes investigation on proteins, DNA’s, drug and pharma industry, bio-informatics etc. The simple and basic of concept of molecular dynamics is to analyse the movements of atoms in the proteins or molecules with the required time step that were being provided which is based on the basic calculations of physics leading interatomic connections [6]. Also, these simulations will provide the extensive variability of important aspects on biological processes such as conformational change, binding I ligands and folding of proteins, visualizing the locations or situations of all the atoms at femtosecond temporal resolution. More-overly this simulation can easily able to analyse the behavior or characters at an atomic level of biomolecules to agitations such as protonation, transformation and adding or removing of ligand [7,8]. The wide range of frequently used molecular dynamics simulation is on techniques on biological experiment structures such as X-ray, crystallography, Cryo-Electron Microscopy (Cryo-EM), Nuclear Magnetic Resonance (NMR) and Electron Paramagnetic Resonance (EPR) (Figure 1).

Figure 1: Simulation of Protein Folding.
In this paper, we have discussed about the necessary for the molecular dynamics simulation in numerous biological features includes, 3 proteins, DNA’s, simulation of drugs and role of the molecular dynamics in biological aspects were discussed.
Role of molecular dynamics in biology
Molecular dynamics simulation is one of the very influential techniques in the advanced molecular biology which allows us to analyse and understand the dynamics and structure in the detailed view accurately on the scales where the movements and motion of the individual atoms can be followed. In both the time and space, the biomolecular dynamics will occur in the extensive series of scales there is a vast difference between the theoretical method and molecular dynamics simulation [9]. The main duty of the molecular dynamics simulation is that to examine whether data of the both theory and experiments were equal or not. For an instance, it is difficult to simulate the ion channel or ions exchange with the experiments but simulation can do it in an easy manner so that it can’t contest with the experiment or either theory, also simulation can provide the detail in depth which is not available with the experiment like pressure dispersals inside the membrane [10]. Both the experimental and theory of the protein structure and the drug designing will improve by the structural refinement and energy minimization which helps to moving towards more exact models are even including complex and large scaled simulations for energy free screening. Rather the properties of any atoms or the molecules can easily be calculated by some equations like Schrodinger equation for time dependent [11].
Though the micro and macroscopic characteristic of the atoms and the molecules can be measured through the experiment it is not the direct observation but it averages over millions of molecules which represents an ensemble of statical mechanics. This is deep calculation which covers in great detail through the theory even practically it includes some mandatory difficulties [12]. Through the experiment it is impossible to work with the individual structures or molecules their behavior but at the conditions of the experiment the ensemble should be generated to expand the system like pressure and temperature. Free energy system such as Binding constants, solubilities and relative stability are the properties based on the thermodynamic equilibrium can’t calculated directly from the molecular dynamics simulations which requires some calculations and some detailed theories [13].
Most commonly used simulations are monte carlo and molecular dynamics simulation. In the molecular dynamics simulation, which are used to produce statistically realistic equilibrium ensembles, though from the simple to complex structure of molecules can give the exact results. The simulation also will face an error are crash when the initial conformation is actually in long from the equilibrium and huge forces which leads to the crash of the simulation or damage the system so that it is mandatory to start with system’s the energy minimization before starting the molecular dynamics simulations [14]. Energy minimization is used to get the structures with the refine very low resolution of the experiments. Every definitive simulation methods depends on the more or less experimental estimates called force fields which are used to calculate the interactions and the systems potential energy to function as the point like coordinates. A potential energy surface is the graph or plot of the mathematical relationship between the molecular structure and the energy [15]. It can able demonstrate the molecules or the atoms having the constant composition, gives where the reaction taken place and describes the relative energies for the consumers. There are some set of the equations of the force field that are used to find the potential energy and some set of parameters were used in the equations. For most cases, these estimates work well, but they cannot duplicate quantum effects such as bond formation or breaking [16]. Molecular dynamics involves some basic steps and procedures to be simulated. This simulation requires some set of parameters based on the type of simulation of molecules. The basic steps involved are initialization, energy minimization, equilibration, characterization, analyzation. In initialization the coordinates should be created, then the initial position of the atoms and molecules is being setup and the velocities of those atoms and molecules are given [17]. Also, the parameters for the calculation of the force field includes potential functions for interactions and particles masses and then the important thing is to setting up the simulation parameters. Parameters are an important thing in the simulation which should be fed properly and correctly with respect to type of the simulation test you go with. Change in parameter or fault in parameter will lead to the error in simulation. So determining the parameter is one of the essential one as it affect the entire result of the simulation. Simulation parameters include step timing, size of the system, and providing ensembles [18]. Afterwards the energy minimization it is nothing but the atomic coordinates of the model that modifies systematically which are resulting in the arrangement of 3-dimensional structure of molecules in the sample, represents the minimized energy or the low resolution is generally named as energy minimization. It is also called geometry optimization. Then in equilibration the forces will be calculated by solving equations like newton’s law of motion and will update the particle position and velocities next, the instantaneous properties like degrees of freedom will be calculated and then finally we can able visualize the final result of motion or movements of the atoms or molecules at the given time step (Figure 2) [19].
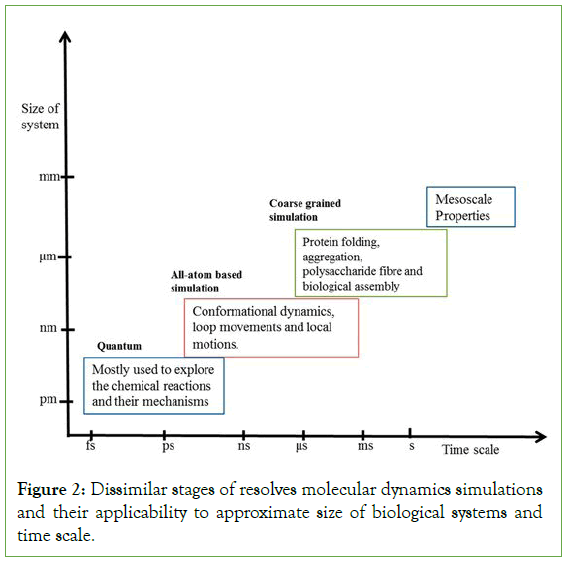
Figure 2: Dissimilar stages of resolves molecular dynamics simulations and their applicability to approximate size of biological systems and time scale.
Advances and advantages molecular dynamics simulation in biology
Understanding and analyzing the theory of science is the eventual aim of the natural singularities [20]. Though, both the experiment and the theory have worked together to understand the results and provide the conclusion in their design, biology is much slower process to squeeze the theoretical approaches in certain cases that are difficult to understand because of some assumptions that are invalid on the experiment and cannot applicable to complex and large systems includes the micro and macro level atomic structures [21]. So, using the computational and analytical approaches those theories were made successful and made their way to biophysics which is rapidly emerging field and significant impact in the application of biological based researchers. Availability of supercomputers have made the new way to development of many sectors includes biology and biological processes. Simulations made the work easier rather than both the theory and the experiment [22]. Biological processes include micro sized atoms that are arranged with millions of molecules that different to identify even at the microscope. Biological micro and macromolecules are not the simple stationary molecules, they are dynamic arrangements and it is not easy to demonstrate every molecule and here there is requirement of simulations to get some accurate results and there is none other option to go with. So molecular dynamics simulation comes into the technology and gives new solution to the many biochemists and biologists [23]. Using the molecular dynamics the movements of the molecules can be visualized by inserting some particular parameters to get the exact results. Molecular dynamics simulation have not main role in molecular motion but also in the modification of macromolecular structures of micro structures based X-ray and space required for free energy evaluation which changes mutation and other perturbations. Molecular dynamics covers over the large number of molecules i.e, from millions of Angstrom to billions of angstrom and a massive range of time-scale from sub-pie seconds to microseconds. Some of these motions are known to have the efficient role [24].
The first step of the molecular dynamics simulation is the choosing the system energy as a function of atomic coordinates. The structure of the different relative stabilities is determined by the potential energy. The forces which are acting on the system that are related with the derivatives with respect to the positions of the atoms which is used to determine the dynamic characteristics of the sample by resolving the newton’s equation of motion for the atoms over the function of time [25]. Though the small molecules require the quantum mechanical calculations, the information of the proteins and the environment is only provided by the functions of the empirical energy. As the protein molecules at the room temperature most of the atom’s motion leaves the length of the bond and the chain of polypeptide angle near the values of equilibrium. The accuracy of the energy functions indicates that the bonding is comparable to the potential energy of small molecules [26].
In newtons equation of motion the detailed information will be provided by molecular dynamics simulation by which it can solve the system of the atoms and molecules and can calculate the for any surrounding solvent. In the simple homogeneous system in which the box is full of water molecules with the required boundary conditions that will provide the structural and dynamic characters can able examine by using the simulations on the few picoseconds [27]. But in non-homogeneous systems such as the proteins, we need to go with extremely larger simulations which require heavy processors and technological computers to run such simulations. Modern computers and computers that have high processors will able to determine and analyse the particles that are even nano-sized allows the simulation to nanoseconds that are adequate to illustrate the liberations pf proteins of the minor groups in proteins [28].
The conventional set of atomic co-ordinates, velocities and forces are obligatory. The coordinates can be found from the structure of the crystals of the X-ray or Nuclear Magnetic Resonance (NVR) or by the building the models like structure of homologous protein. Next the velocities and the forces of the atoms were allocated from Maxwellian distribution at a very low temperature and in the few pie-seconds the simulations were being performed [29]. By using the newton’s second law of motion, the acceleration of the atom can be obtained, F=ma where a is the acceleration of the molecules and F defines the force of the atoms which is calculated from motion and derivatives regarding the position and m is the mass. The position r, at time t

Alternating the innovative velocity which is continued by the Maxwellian distributions at different temperatures. By which the temperature of the system is measured by the mean kinetic energy.
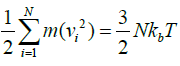
Where N defines the number of atoms and v is the average velocity of the atom and Kb is the Boltzmann constant [30].
Application of molecular dynamics in biological aspects
Molecular dynamics simulation in proteins: Proteins are called as the building blocks of amino acids and mainstays of biological systems which achieve most of the tasks essential to maintain the life. Though the mechanisms of protein folding have been studied over the years by the means of both the experimental and simulation, the determination of the protein sequence, the ordering of amino acids, its alignment and arrangement which specifies the structure and function is very challenging in the day-to-day life of molecular biology [31]. The study was made on. The simulation of the protein molecules is being programmed in the structure of the molecules and is always a serious component of their function. It is essential to understand and analyzing the protein molecules through molecular dynamics simulation and the connection between the three-dimensional structures which are obtained with increasing rapidity by Nuclear Magnetic Resonance (NMR) and X-ray that are very much difficult to investigate experimentally [32]. Simulation through molecular dynamics also provides the connection between the dynamics and structure by permitting the examination of the movement of molecules, where the protein molecules are accessible by the conformational energy landscape [33]. In 1977, the first simulation on proteins using the molecular dynamics was reported and it consists of 9.2-ps, route to a minute protein particle in vacuum. Similarly, after some years the same protein on 210-ps simulation in water has been reported and which leads to the remarkable rise in calculating power as now simulations were carried out for large proteins that are thousand times long as that of the original simulation, in which the same protein is enclosed by salt and water [34]. Molecular dynamics simulation provides abundant dynamical structural data on biomolecules and also energetic data on molecules of proteins and interaction of ligands. In the function, the substantial developments were also been attained that makes simulation steadier and more precise. Simulations give more effective results when it is being analysed in closed combination while compared to the experiments that plays the vital role over validating and improving the simulations [35]. In recent, simulation of comprehensive folding routes in explicit solvent is not possible if the long time-scales essential for process of folding have destined. Also, the simulations of the protein folding had usually complete use of coarse models, implied solvent, or for the sort trajectories of very large ensembles were used to get info on the physical folding pathway [36].
In every case, the accuracy for the test of the structural mechanism is mandatory or even to refine it. i.e, because of the packing of the crystal lattice will lead to the defects in the crystal structure and even for the protein molecules due to the absence of lipid layer. So there is the requirement of numerical simulations because, those defects can easily be corrected by preliminary from the crystal structure, then in an suitable incorporate atmosphere and permitting the structure to diminish to a more favorable conformation [37]. Also, molecular dynamics are used to recuperate the ensembles of conformations as opposed to single structure. Molecular dynamics simulations will also provide molecular structures which from very low resolution of Cryo-Electron microscope maps to the complex structure of high resolution is also be provided which are based on some protocols. Cryo-EM has become more advanced in structural modelling which being examined by Eman et al [38].
The requirement of the molecular dynamics simulation is higher due to its advancement and more efficient for the persons who is working with Molecular Dynamics (MD) as they give proper results, to build or improve the structural models which are based on the data’s of the biological structures and crystal structures of X-ray [39- 41]. Particularly molecular dynamics simulation is used in the main application to regulate how the system of biomolecules will react to some agitation. Dror et al. experimented the structure of proteins to remove the bound ligand and they simulated to visualize that the protein conformation is affected when ligand is removed [42,43]. If the bound ligand is replaced by some other ligands or adding ligands so that the none of them were present in the experimental structure. The mutation of one or more amino acids in the protein can able to visualize their reactions in different cases going with the MD simulation rather going with the experiment [44,45].
The main purpose for the molecular dynamics simulation is to analyse the action of biomolecular processes in which the particular processes such as protein folding, transport of membrane, ligand binding, ligand-induced conformational change. These allow us to resolve various problems that are difficult while experimented. Through the molecular dynamics simulation, it can be made easier and every phases and movement of the single amino-acids of proteins (Figure 3) [46,47].
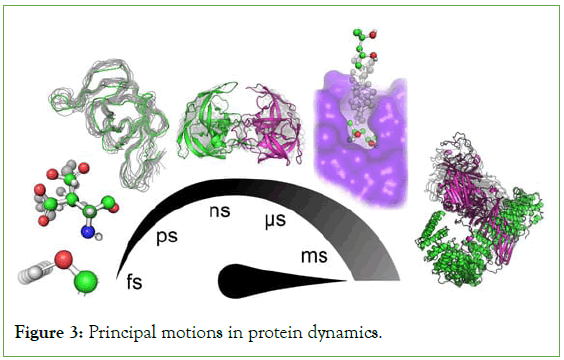
Figure 3: Principal motions in protein dynamics.
Molecular dynamics simulations have a great impact on experiment testing by the statistic that the most of the standards of Molecular dynamics lies in the investigation of the properties of molecules or the cells which are problematic or unbearable to go with the experiments. In some aspects, the tests were carried out through the simulations as they simply reasonable such as ligands and the design of the proteins, though through the experiments, binding energy filtration or winnow down a large pool of candidates can be investigated [49-50]. But still, in biological aspects such as understanding the of biomolecules and working of drugs, in those cases because of its limitations, there is an unavailability of experiment by which it could afford the same results while being simulated. Though the results taken from the experiment were designed to get specific calculations and results taken from the simulations generally authenticate the simulation results. Conceivably even more importantly, simulations will also make premises which lead to new experimental work [51].
Molecular dynamics simulations in drugs: Molecular dynamics simulations consume a significant role in the expansion process of modern drug and chemical industries. It becomes more advance in the growth of pharmaceutical industry [52]. The main use of the MD in those applications are to analyse the function of the proteins present in the drugs and to identify the role of the antibody design such as RAS proteins, essentially disordered proteins is their main duty. The different phases that involve during the discovery of the drug are several identifications of the target and validating, detection of lead compounds, optimization of lead, research of the particular drug, trials of the particular drugs for the previous three phases and then finally into the market with authorization [53]. The different phases includes the different distinctive movements, approaches, or tools used in the specific development phase which includes bio-chemistry, bio-informatics. The different phases includes the different distinctive actions, methods, or tools used in the particular development phase which includes bio-chemistry, bio-informatics, biochemistry, proteomics were used [54]. For the discovery for the lead compounds natural substances, compound libraries were used, in the optimization of lead chem-informatics, molecular modelling were used, neatly based on the research the simulation of the particular drug under some certain conditions is carried out through molecular dynamics simulation [55]. There are thousands of molecules present in drug that cannot be visualized or difficult in experiment so by using the molecular dynamics the movement of particles or the reaction of the particles over the period of time can be visualize the specific atoms or the group of segments at the same time [56]. The reaction or the behavior of the protein molecules are very essential in every cases in the molecular dynamics simulation, using this we can conclude the result if there is any change in simulation it can also be altered to get complete and the exact results [57]. For an example, in several drug and chemical industries they will simulate the molecules includes proteins, chemicals etc. to calculate the where reaction takes place and when the reaction occurs and reason for the result and drug is an important one which is linked to health of the human beings. In drug industry for analyzing the validation of the drug they will go with MD simulation to get the exact result of the expiry date. So the molecular dynamics is important in every sectors including in the drug industries (Figure 4) [58-60].
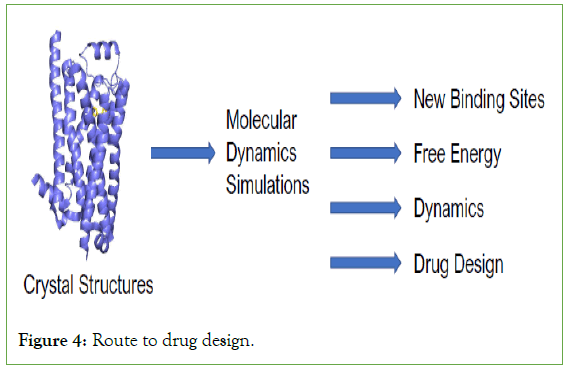
Figure 4: Route to drug design.
Prediction of the structure is one of most antique problems which were addressed in the structural bioinformatics. The large simulations of complex structures can also be simulated using the Molecular dynamics and it is used for prediction of an initio protein structure, pointing to simulate the folding of protein from scratch, though it is not an actual approach to attain the theoretical model for the structure of proteins (Figure 5) [61].

Figure 5: Different phases involved in molecular dynamics simulations of drug discovery.
Role of molecular dynamics simulations assist with gene medicines: By using the molecular dynamics simulation, it provides an ability to see complex atomical and molecular structures at the boundary of the biomaterials and Biosystems [62,63]. Hasan et al. made his investigation on role of molecular dynamics of gene and how it assists gene medicines [64]. And using the molecular dynamics simulation, Gene therapy exertions that organize biomaterial intermediated distribution of nucleic acids could benefit immensely. These effort leads to the final results of supramolecular assembly having structures of very high dynamics and essential [65]. The characteristics behavior of the elements can be easily visualized by careful analysis of MD simulation and gives the way to biosystems. The recently developed gene therapy is an essential part of biomaterials have made the possible efforts to understand the clinical advantage of nucleic-acids [66].
Molecular dynamics simulations interactions of DNA and DNAprotein: The interactions of molecular level in DNA and DNAprotein systems can easily be characterized for every atom by using the molecular dynamics simulation with extraordinary resolution [67]. The technologies of computational analysis have become more advance in the recent years and permitted to visualize those impartial characteristics behavior of the system even at the timescale of micro-second while the sample which is enhanced or improved the sample finds its way understand deeply the characterization of free energy interaction and its interactions with the numerical accuracy [68]. Jejoong examined the simulations increases the realism in the recent progress by cleansing the accuracy of the MD force field.
Simulation of the heterogenous cells through MD simulation: The signatures of diseased cells of the chemo-biomechanical are always characteristically varies from that of the healthy cells. These cells are generally generated from the alterations of the cellular structural persuaded by the development of disease and beneficial molecules. Therapeutic shock waves have the ability to kill the cells automatically that are affected or increase the permeability of the cell membrane for the process of the drug delivery [69,70]. Still, through which the interaction of shock waves with the affected as well as healthy cellular components of the biomolecular mechanism endures largely unidentified. With the help of the molecular dynamic simulations, it is easy to understand and analyse the characteristics of the biomolecules at the multi scaled numerical framework through which it offers the perspectives [71]. Here they inspect the responses of the biomechanical sectors that have the characteristic of complexes membrane below quick mechanical loadings pertinent to therapeutic shock wave conditions. Lili et al. investigated their study on effect of the strain rate which determines the characteristics of rupture which doesn’t display substantial sensitivity [72]. Also, they showed that under the applied membrane stretches, the embedded rigid inclusions distinctly enable stretchinduced membrane interruptions while instinctively hardening the related developments. They conclude that the due to the occurrence of the rigid molecules in the membranes of the cellular which also assists as the mechanical catalyst that endorse as mechanical obliterations of the allied developments, also they concert with the bio-chemical and medical applications which offers the valuable instructions and advancement in the biomechanical-mediated therapeutics which is shown is Figure 6 [73].
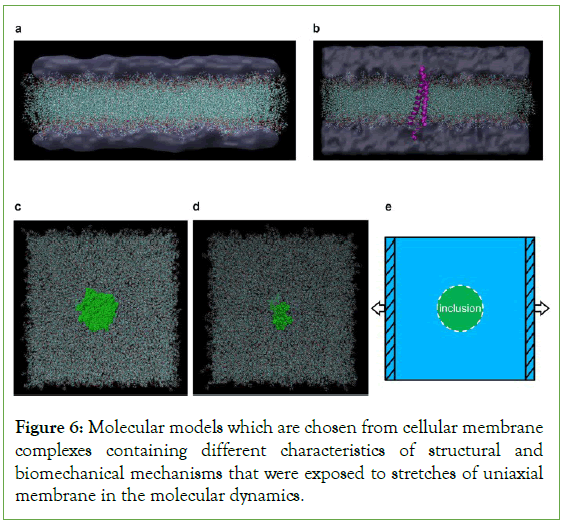
Figure 6: Molecular models which are chosen from cellular membrane complexes containing different characteristics of structural and biomechanical mechanisms that were exposed to stretches of uniaxial membrane in the molecular dynamics.
Conclusion
Thus, molecular dynamics simulation becomes more advanced and becomes an essential role in every sector especially in biology. Simulation reduces the time and gives the exact result. Biological experiments deal with the identifying and analyzing the biomolecular structures includes movement of the individual molecules over the period of time. Biological aspects include proteins, DNA, bioinformatics, drugs, chemicals, medicines and even in the food processing. Biomolecular structures include cells of nano and micro sized cells and even in angstrom. Molecular dynamics is the advance technology dealing with the simulating the atoms/molecules movements of the system, when applied to biological micro-molecules which gives the variation in the different locations of the cells in the proteins and in DNS over the function of time. Thus molecular dynamics is such a developed science which deals with the interaction in between the molecules. Molecular dynamics is not simple antique spectacle, but it is quickly emerging field of science especially in the field of biological aspects that includes investigation on proteins, DNA’s, Drug and pharma industry, bio-informatics etc. The simple and basic of concept of molecular dynamics is to analyse the movements of atoms in the proteins or molecules with the required time step that were being provided which is based on the basic calculations of physics leading interatomic connections. In both the time and space the biomolecular dynamics will occur in the extensive series of scales there is a vast difference between the theoretical method and molecular dynamics simulation [74].
References
- Gupta S, Varadwaj PK. A brief overview on molecular dynamics simulation of biomolecular system: Procedure, algorithms and applications. Int J Pharm Sci Res. 2018; 9(4):1333-1350.
- Yao Z, Zhu CC, Cheng M, Liu J. Mechanical properties of carbon nanotube by molecular dynamics simulation. Comput Mater Sci. 2001; 22(3-4):180-184.
- Shen X, Bourg IC. Molecular dynamics simulations of the colloidal interaction between smectite clay nanoparticles in liquid water. J Colloid Interface Sci. 2021; 584:610-621.
[Crossref] [Google Scholar] [PubMed]
- Boldon L, Laliberte F, Liu L. Review of the fundamental theories behind small angle X-ray scattering, molecular dynamics simulations, and relevant integrated application. Nano Rev. 2015; 6(1):25661.
[Crossref] [Google Scholar] [PubMed]
- Neves RP, Sousa SF, Fernandes PA, Ramos MJ. Parameters for molecular dynamics simulations of manganese-containing metalloproteins. J Chem Theory Comput. 2013; 9(6):2718-2732.
[Crossref] [Google Scholar] [PubMed]
- Koopman EA, Lowe CP. Advantages of a Lowe-Andersen thermostat in molecular dynamics simulations. J Chem Phys. 2006; 124(20):204103.
[Crossref] [Google Scholar] [PubMed]
- Teodoro ML, Phillips Jr GN, Kavraki LE. Understanding protein flexibility through dimensionality reduction. J Comput Biol. 2003; 10(3-4):617-634.
[Crossref] [Google Scholar] [PubMed]
- Kannan S, Zacharias M. Folding of Trp-cage mini protein using temperature and biasing potential replica-exchange molecular dynamics simulations. Int J Mol Sci. 2009;10(3):1121-1137.
[Crossref] [Google Scholar] [PubMed]
- Lindahl ER. Molecular dynamics simulations. J Mol Model. 2008:3-23.
- Paquet E, Viktor HL. Molecular dynamics, Monte Carlo simulations, and langevin dynamics: A computational review. Biomed Res Int.2015.
[Crossref] [Google Scholar] [PubMed]
- Feig M. Computational protein structure refinement: Almost there, yet still so far to go. Wiley Interdiscip Rev Comput Mol Sci. 2017; 7(3):e1307.
[Crossref] [Google Scholar] [PubMed]
- Lebowitz, JL. Microscopic reversibility and macroscopic behaviour: Physical explanations and mathematical derivations.
- Van Gunsteren WF, Berendsen HJ. Computer simulation of molecular dynamics: Methodology, applications, and perspectives in chemistry. Angew Chem Int Ed Engl. 1990; 29(9):992-1023.
- Grossfield A, Patrone PN, Roe DR, Schultz AJ, Siderius DW, Zuckerman DM. Best practices for quantification of uncertainty and sampling quality in molecular simulations .Living J Comput Mol Sci. 2018; 1(1).
[Crossref] [Google Scholar] [PubMed]
- Szeliski R, Zabih R, Scharstein D, Veksler O, Kolmogorov V, Agarwala A, et al. A comparative study of energy minimization methods for markov random fields.
- Boykov Y, Veksler O, Zabih R. Fast approximate energy minimization via graph cuts. IEEE Trans Pattern Anal Mach Intell. 2001; 23(11):1222-1239.
- Wereszczynski J, McCammon JA. Statistical mechanics and molecular dynamics in evaluating thermodynamic properties of biomolecular recognition. Q Rev Biophys. 2012; 45(1):1-25.
[Crossref] [Google Scholar] [PubMed]
- Vanommeslaeghe K, Guvench O. Molecular mechanics. Curr Pharm Des. 2014; 20(20):3281-3292.
[Crossref] [Google Scholar] [PubMed]
- Zacharias M. ATTRACT: Protein–protein docking in CAPRI using a reduced protein model. Proteins: Struct Funct and Bioinform. 2005 Aug; 60(2):252-256.
[Crossref] [Google Scholar] [PubMed]
- Singh N, Li W. Recent advances in coarse-grained models for biomolecules and their applications. Int J Mol Sci. 2019; 20(15):3774.
[Crossref] [Google Scholar] [PubMed]
- Bernardi RC, Melo MC, Schulten K. Enhanced sampling techniques in molecular dynamics simulations of biological systems. Biochim Biophys Acta. 2015; 1850(5):872-877.
[Crossref] [Google Scholar] [PubMed]
- Wooley JC, Lin HS. Computational modelling and simulation as enablers for biological discovery. 2005.
- Gennis RB, editor. Biomembranes: Molecular structure and function. 2013.
- Yang W, Pan Y, Zheng F, Cho H, Tai HH, Zhan CG. Free-energy perturbation simulation on transition states and redesign of butyrylcholinesterase. Biophys J. 2009; 96(5):1931-1938.
[Crossref] [Google Scholar] [PubMed]
- Levitt M, Hirshberg M, Sharon R, Daggett V. Potential energy function and parameters for simulations of the molecular dynamics of proteins and nucleic acids in solution. Comput Phys Commun. 1995;91(1-3):215-231.
- MacKerell Jr AD, Bashford D, Bellott ML, Dunbrack Jr RL, Evanseck JD, Field MJ, et.al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998; 102(18):3586-3616.
[Crossref] [Google Scholar] [PubMed]
- Karplus M. Molecular dynamics of biological macromolecules: A brief history and perspective. Biopolymers. 2003; 68(3):350-358.
[Crossref] [Google Scholar] [PubMed]
- Katiyar RS, Jha PK. Molecular simulations in drug delivery: Opportunities and challenges. Wiley Interdiscip Rev Comput Mol Sci. 2018; 8(4):e1358.
- Hyperpolarisation CS, SPonSorS P, Hall HSOR, Hall PLOR, and Theatre, L poster presentations-euromar 2012.
- Volz SG, Chen G. Molecular-dynamics simulation of thermal conductivity of silicon crystals. Phys Rev B. 2000; 61(4):2651.
- Patodia S, Bagaria A, Chopra D. Molecular dynamics simulation of proteins: A brief overview. J Phys Chem Biophys. 2014;4(6):1.
- Adcock SA, McCammon JA. Molecular dynamics: Survey of methods for simulating the activity of proteins. Chem Rev. 2006;106(5):1589-1615.
[Crossref] [Google Scholar] [PubMed]
- Borhani DW, Shaw DE. The future of molecular dynamics simulations in drug discovery. J Comput Aided Mol Des. 2012;26:15-26.
[Crossref] [Google Scholar] [PubMed]
- Karplus M, Kuriyan J. Molecular dynamics and protein function. Proc Natl Acad Sci USA. 2005;102(19):6679-6685.
[Crossref] [Google Scholar] [PubMed]
- Grossfield A, Feller SE, Pitman MC. Convergence of molecular dynamics simulations of membrane proteins. Proteins. 2007;67(1):31-40.
[Crossref] [Google Scholar] [PubMed]
- Juraszek J, Bolhuis PG. Sampling the multiple folding mechanisms of Trp-cage in explicit solvent. Proc Natl Acad Sci USA. 2006;103(43):15859-15864.
[Crossref] [Google Scholar] [PubMed]
- Kuzmanic A, Pannu NS, Zagrovic B. X-ray refinement significantly underestimates the level of microscopic heterogeneity in biomolecular crystals. Nat Commun. 2014;5(1):3220.
[Crossref] [Google Scholar] [PubMed]
- Singharoy A, Teo I, McGreevy R, Stone JE, Zhao J, Schulten K, et al. Molecular dynamics-based refinement and validation for sub-5 Å cryo-electron microscopy maps. Elife. 2016;5:e16105.
[Crossref] [Google Scholar] [PubMed]
- Gagnon E, Ranitovic P, Tong XM, Cocke CL, Murnane MM, Kapteyn HC, et al. Soft X-ray-driven femtosecond molecular dynamics. Science. 2007;317(5843):1374-1378.
[Google Scholar] [PubMed]
- Brunger AT, Adams PD. Molecular dynamics applied to X-ray structure refinement. Acc Chem Res. 2002;35(6):404-412.
[Crossref] [Google Scholar] [PubMed]
- Hura G, Russo D, Glaeser RM, Head-Gordon T, Krack M, Parrinello M, et al. Water structure as a function of temperature from X-ray scattering experiments and ab initio molecular dynamics. Phys Chem Chem Phys. 2003;5(10):1981-1991.
- Hospital A, Goñi JR, Orozco M, Gelpí JL. Molecular dynamics simulations: Advances and applications. Adv Appl Bioinform Chem. 2015;19:37-47.
[Crossref] [Google Scholar] [PubMed]
- Sagui C, Darden TA. Molecular dynamics simulations of biomolecules: Long-range electrostatic effects. Adv Appl Bioinform Chem. 1999;28(1):155-179.
[Crossref] [Google Scholar] [PubMed]
- Mobley DL, Dill KA. Binding of small-molecule ligands to proteins: “What you see” is not always “what you get”. Structure. 2009;17(4):489-498.
[Crossref] [Google Scholar] [PubMed]
- Naqvi AA, Mohammad T, Hasan GM, Hassan M. Advancements in docking and molecular dynamics simulations towards ligand-receptor interactions and structure-function relationships. Curr Top Med Chem.2018; 18(20):1755-1768.
[Crossref] [Google Scholar] [PubMed]
- Mori T, Miyashita N, Im W, Feig M, Sugita Y. Molecular dynamics simulations of biological membranes and membrane proteins using enhanced conformational sampling algorithms. Biochim Biophys Acta. 2016;1858(7):1635-1651.
[Crossref] [Google Scholar] [PubMed]
- Shukla R, Tripathi T. Molecular dynamics simulation of protein and protein-ligand complexes. Curr Comput Aided Drug Des. 2020:133-161.
- Souza PC, Thallmair S, Conflitti P, Ramírez-Palacios C, Alessandri R, Raniolo S, et al. Protein-ligand binding with the coarse-grained martini model. Nat Commun. 2020;11(1):3714.
[Crossref] [Google Scholar] [PubMed]
- Scheraga HA, Khalili M, Liwo A. Protein-folding dynamics: Overview of molecular simulation techniques. Annu Rev Phys Chem. 2007;58:57-83.
[Crossref] [Google Scholar] [PubMed]
- Di Nola A, Roccatano D, Berendsen HJ. Molecular dynamics simulation of the docking of substrates to proteins. Proteins.1994;19(3):174-182.
[Crossref] [Google Scholar] [PubMed]
- Salsbury Jr FR. Molecular dynamics simulations of protein dynamics and their relevance to drug discovery. Curr Opin Pharmacol. 2010;10(6):738-744.
[Crossref] [Google Scholar] [PubMed]
- Durrant JD, McCammon JA. Molecular dynamics simulations and drug discovery. BMC biol. 2011;9(1):1-9.
[Crossref] [Google Scholar] [PubMed]
- John Fox S, Li J, Tan YS, N Nguyen M, Pal A, Ouaray Z, et al. The multifaceted roles of molecular dynamics simulations in drug discovery. Curr Pharm Des. 2016;22(23):3585-3600.
[Crossref] [Google Scholar] [PubMed]
- Liu X, Shi D, Zhou S, Liu H, Liu H, Yao X, et al. Molecular dynamics simulations and novel drug discovery. Expert Opin Drug Discov. 2018;13(1):23-37.
[Crossref] [Google Scholar] [PubMed]
- Bera I, Payghan PV. Use of molecular dynamics simulations in structure-based drug discovery. Curr Pharm Des. 2019;25(31):3339-3349.
[Crossref] [Google Scholar] [PubMed]
- Salo-Ahen OM, Alanko I, Bhadane R, Bonvin AM, Honorato RV, Hossain S, et al. Molecular dynamics simulations in drug discovery and pharmaceutical development. Processes. 2020;9(1):71.
- Borhani DW, Shaw DE. The future of molecular dynamics simulations in drug discovery. J Comput Aided Mol Des. 2012;26:15-26.
[Crossref] [Google Scholar] [PubMed]
- Kerrigan JE. Molecular dynamics simulations in drug design. Methods Mol Biol. 2013:95-113.
[Crossref] [Google Scholar] [PubMed]
- Weng L, Stott SL, Toner M. Exploring dynamics and structure of biomolecules, cryoprotectants, and water using molecular dynamics simulations: Implications for biostabilization and biopreservation. Annu Rev Biomed Eng. 2019;21:1-3.
- Shariatinia Z. Molecular dynamics simulations on drug delivery systems. 2021
- Jothi A. Principles, challenges and advances in ab initio protein structure prediction. Protein Pept Lett. 2012;19(11):1194-1204.
[Crossref] [Google Scholar] [PubMed]
- Zou Y, Ewalt J, Ng HL. Recent insights from molecular dynamics simulations for g protein-coupled receptor drug discovery. Int J Mol Sci. 2019;20(17):4237.
[Crossref] [Google Scholar] [PubMed]
- Salo-Ahen OM, Alanko I, Bhadane R, Bonvin AM, Honorato RV, Hossain S, et al. Molecular dynamics simulations in drug discovery and pharmaceutical development. Processes. 2020;9(1):71.
- Uludağ H, Tang T. How can molecular dynamics simulations assist with gene medicines? Biomater Biosyst. 2021;2:100014.
[Crossref] [Google Scholar] [PubMed]
- Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregersen BA, et al. Scalable algorithms for molecular dynamics simulations on commodity clusters.2006.
- Perilla JR, Goh BC, Cassidy CK, Liu B, Bernardi RC, Rudack T, et al. Molecular dynamics simulations of large macromolecular complexes. Curr Opin Struct Biol. 2015;31:64-74.
[Crossref] [Google Scholar] [PubMed]
- Yoo J, Winogradoff D, Aksimentiev A. Molecular dynamics simulations of DNA–DNA and DNA–protein interactions. Curr Opin Struct Biol. 2020;64:88-96.
[Crossref] [Google Scholar] [PubMed]
- Matouš K, Geers MG, Kouznetsova VG, Gillman A. A review of predictive nonlinear theories for multiscale modelling of heterogeneous materials. J Comput Phys. 2017;330:192-220.
- Lyman E, Pfaendtner J, Voth GA. Systematic multiscale parameterization of heterogeneous elastic network models of proteins. Biophys J. 2008;95(9):4183-4192.
[Crossref] [Google Scholar] [PubMed]
- Hoteit H, Firoozabadi A. Numerical modeling of two-phase flow in heterogeneous permeable media with different capillarity pressures. Adv Water Resour. 2008;31(1):56-73.
- Reuter K, Scheffler M. First-principles kinetic Monte Carlo simulations for heterogeneous catalysis: Application to the CO oxidation at RuO2 (110). Phys Rev B. 2006;73(4):045433.
- Zhu H, Kee RJ, Janardhanan VM, Deutschmann O, Goodwin DG. Modelling elementary heterogeneous chemistry and electrochemistry in solid-oxide fuel cells. J Electrochem Soc. 2005;152(12):A2427.
- Smith GD, Bedrov D, Li L, Byutner O. A molecular dynamics simulation study of the viscoelastic properties of polymer nanocomposites. Chem Phys. 2002;117(20):9478-9489.
- Zhang L, Zhang Z, Jasa J, Li D, Cleveland RO, Negahban M, et al. Molecular dynamics simulations of heterogeneous cell membranes in response to uniaxial membrane stretches at high loading rates. Sci Rep. 2017;7(1):8316.
[Crossref] [Google Scholar] [PubMed]
Citation: Kannan S (2023) A Review on Molecular Dynamics Simulation in Nano and Micro-sized Biomolecular Structures in Biological Aspects: Its Advancement and Advances. Biochem Anal Biochem. 12:492.
Copyright: © 2023 Kannan S. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.


