Indexed In
- ResearchBible
- CiteFactor
- RefSeek
- Hamdard University
- EBSCO A-Z
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Original Research Article - (2023) Volume 12, Issue 4
Use of Media Conditioning for Robust Single Cell Sorting of Induced Pluripotent Stem Cells
Christophe Michel Raynaud* and Sharefa Al-MannaiReceived: 02-Oct-2023, Manuscript No. SCPM-23-23214; Editor assigned: 06-Oct-2023, Pre QC No. SCPM-23-23214 (PQ); Reviewed: 20-Oct-2023, QC No. SCPM-23-23214; Revised: 27-Oct-2023, Manuscript No. SCPM-23-23214 (R); Published: 04-Nov-2023, DOI: 10.35248/2168-9431.23.12.068
Abstract
Human Pluripotent Stem Cells (hPSCs) constitute a unique tool for disease modeling and functional genomics. Such applications require the generation of cell clones established by repetitive cycles of tissue growth, dissociation, and re-seeding. These cloning cycles are time-consuming and increase the likelihood of genetic instability. We defined feeder-free culture conditions on Matrigel in Stem-Flex medium to establish a single-cell cloning workflow using Fluorescent Activated Cell Sorting (FACS). The workflow relies on the Stem-Flex medium supplemented with a conditioned medium and allows the efficient cloning of newly reprogramed and established human induced pluripotent stem cells lines. This workflow is an efficient and cost-effective method for the rapid derivation of stem cell clones after reprogramming or gene editing.
Keywords
Human pluripotent stem cells; Single-cell sorting; Conditioned media
Introduction
Human Pluripotent Stem Cells (hPSCs), either of Human Embryonic Origin Cells (hESCs) or reprogrammed from mature/ differentiated human adult cells into induced Pluripotent Stem Cells (hiPSCs), are of great interest for research and clinical applications [1]. The generation and differentiation of patient- derived hiPSCs are one of few approaches for modeling diseases of cells that would otherwise be difficult or impossible to isolate from patients, such as neurons or cardiomyocytes. Despite tremendous progress in reprogramming cells with numerous non-integrative, feeder-free methods, the labor-intensive cloning process of deriving iPSCs is the limiting step [2]. The expansion of hPSCs relies on passaging cells as “small colonies” or “clumps” to maintain their undifferentiated state, avoid cell death, and prevent the selection of subpopulations [3]. In addition to being a disease model of inaccessible differentiated cells, hiPSCs and hESCs are at the center of disease modeling by personalized gene editing with systems such as Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) and CRISPR associated 9 (Cas9) endonuclease. Genome editing allows the reversion, or introduction, of mutations suspected to be responsible for a given phenotype while keeping the remainder of the genomic background unaffected, therefore, allowing the isolation and the investigation of the physiological consequences of a given mutation. In “normal” hiPSCs, the introduction of mutations potentially associated with a disease removes the need for systematically deriving hiPSCs from affected patients. Finally, genome editing can be used to tag endogenous proteins with reporter genes, creating lines [4], which maintain the physiological expression of the tagged proteins under their endogenous promoter. In any case, these gene-editing systems require the cloning and the expansion of modified clones. This process rapidly becomes tedious and extremely expensive depending on the CRISPR/Cas9 edition efficiency.
Single-cell dissociation of Human Pluripotent Stem Cells (hPSCs) drives genomic instability and drastically affects cell viability [5]. The use of Rho-Associated Protein Kinase (ROCK) inhibitors, a cocktail of small molecules (small molecule cocktail of 4 inhibitors: SMC4 cocktail), and rapamycin mitigate this problem [6,7]. In most cases, repetitive manual colony picking and re-expansion over long periods are performed, and these solutions remain inefficient in some cell lines [8]. Newly developed culture methods, media, and matrixes have improved single-cell viability in culture [6,9]. Specifically, in the last few years, introducing “stabilized” βFGF has done away with the need for daily spiking of culture media with βFGF to maintain the exposition of hPSCs to this factor. These new stabilized media allow for a more stable and constant cell-culture condition [10]. The method of passage in small clumps highlights the need for a “herd effect”, reflecting the necessity of cell-cell interactions and the exchange of soluble factors such as cytokines and Microvesicles (MVs) for cell survival and stability.
MVs are lipid vesicles with a diameter ranging from 30 nm to 1 uM. They are released by all cell types and contain microRNA, mRNA, protein, and lipids. hPSCs are no exception; they release MVs [11,12].
An alternative to the repetition of the culture cycle expansion- dissociation-reseeding to clone stem cells after many cycles would be the physical sorting and plating of individual hPSCs. Unfortunately, the poor resistance to mechanical stress and isolation remains a barrier for the single-cell sorting of hPSCs. A recent publication demonstrated that iPSCs could be sorted with acceptable survival while maintaining pluripotency and genomic integrity using a newly developed matrix and medium [4,13]. This approach, demonstrated on well-established cell lines (passage 20- 30), has a variable efficiency between cell lines.
In this study, we minimized the cell-sorting-induced mechanical stress by using a large-bore sorting nozzle. Additionally, we “mimicked” the herd effect using hPSCs “conditioned media” or purified MVs. Combining both elements established a reliable and efficient single cell sorting method of newly reprogrammed hiPSCs and well-established cell lines. We demonstrate that media conditioned by hPSCs can be used in combination with ROCK inhibitor to enable the survival of single cell sorted hPSCs. This phenomenon seems to depend on soluble factors that do not involve MVs.
Materials and Methods
Human pluripotent stem cells and culture
We obtained Embryonic stem cell lines ES4 and H1 hESCs from WiCell, and the Human Episomal iPSC (HE iPSC) line from Thermofisher scientific (#A18945). We generated other iPSCs as described below. We maintained all hPSCs on Matrigel (Corning) coated plate in Stem-Flex medium (ThermoFisher). Plates were coated with BD Matrigel growth factor reduced (#354230) by diluting Matrigel with cold (4°C) StemFlex media (1 in 24 ml) and dispensing 2 ml per well of a 6-well plate or 200 μl per well of 96-well plate. Matrigel was incubated overnight at 4°C prior to polymerization at 37°C for 60 minutes. We rinsed excess Matrigel with fresh prewarmed (37°C) media. We changed media every day except otherwise stated. Cells were released from the culture dishes by treatment with ReLesR (Stemcell technologies) for 3 min at 37°C. After ReLesR aspiration, cells clumps were further dissociated using 1 ml of StemFlex media with a 1 ml pipette and subsequently re-seeded on new Matrigel-coated plates. We carried out all cultures at 37°C, 5% CO2, 95% humidity.
When indicated, we used Y-27632 (rock inhibitor) (R&D system,#1254/1) at a final concentration of 10 μM, StemBoostTM Reprogramming Cocktail, referred to as SMC4 cocktail (Biovision,#S231) and Revitacell supplement (Termofisher,#A2644501) at final dilution 1:100 according to manufacturer recommendation.
IPS reprogramming
Blood samples were obtained under the IRB protocol#1702007608 and#1702007592 of Sidra Medicine or purchased from StemCell technologies (#70025.1). Peripheral mononucleated cells (PBMCs) were isolated using Ficoll-paque (GE healthcare) as previously described [14]. hiPSCs were derived from whole PBMCs by transduction with Sendai viruses provided in the CytoTune iPS 2.0 Sendai Reprogramming Kit according to manufacturer recommendation, using StemFlex medium instead of E8 medium. Briefly, PBMCs were cultured in StemPro 34 complemented with SCF (100 ng/ml), FLT-3 (100 ng/ml), IL-3 (20 ng/ml) and IL-6 (20 ng/ml) for four days prior to transduction. Three days after transduction, cells were passed on Matrigel-coated dishes and cultured in media without cytokines. At day 7 post-transduction, the medium was changed progressively to StemFlex. The following culture was performed as described above for hPSCs.
hPSCs staining and single-cell sorting
We performed surface marker analysis with the following antibodies: SSEA4 conjugated with PhycoErythrin (Biolegend) and TRA-1-60 conjugated with Alexa Fluor 488 (Invitrogen). These two markers are the best surface marker for selecting reprogrammed hIPSCs [15,16]. While non-fully reprogrammed stem cells can express Tra1- 60 on their surface in the early stages of reprogramming, Tra1-60 is the most robust marker of pluripotency from day 21 post-reprogramming [15,17]. Fluorescent activated cell sorting (FACS) was performed on a BD aria III cell sorter (Beckton Dickinson) as previously published to accommodate hPSCs.
Briefly, we dissociated cells with ReLesR (Stemcell technologies) for 5 min. After ReLesR aspiration, we further dissociated cell clumps with 1 ml of Dulbecco’s phosphate-buffered saline (DPBS) without Ca2+ and Mg2+ with a 1 ml pipette. Cells were counted using a TC20 automated cell counter (Biorad). Cells were pelleted by centrifugation at 120 g for 4 min with low brake. Pellets were resuspended in 100 ul of DPBS without Ca2+ and Mg2+ and stained with Zombie UVTM Fixable Viability dye (Biolegend) according to manufacturer recommendation (1 ul per million cells) followed by staining at room temperature for 15 min with TRA-1-60 AF488 (dilution 1:50) and SSEA4-PE (dilution1:20). Cells were then rinsed in 6 ml DPBS without Ca2+ and Mg2+ with centrifugation at 120 g for 4 min with low brake. Cells were then resuspended in StemFlex Media prior to filtration on 40 μm mesh (Becton Dickinson#352235). Cell sorting was performed immediately. Data were acquired and processed with FACSDiva 6.3 software (BD Biosciences). We used a 100 μm nozzle to sort the cells at a sheath pressure of 20 PSI. We established an initial gate on forward and side light scatter (SSC-A × FSC-A), we excluded doublets by forward and side scatter height vs. width analysis (FSC-W × FSC-H and SSC-W × SSC-H). We excluded dead cells based on their positive staining with Zombie UV Viability dye (excitation 355 nm, filter 450/50). AlexaFluor 488 (AF488) fluorescence was acquired with 488 nm blue laser excitation and a 505 long-pass filter followed by a 525/50 nm bandpass filter (FITC). Phycoerythrine (PE) was acquired with a 514 nm green laser excitation and a 586/15 nm filter emission (PE). Positivity was defined based on unstained and Fluorescence Minus One (FMO) samples. Newly reprogrammed hiPSCs gating was defined based on the antigen expression level of an established cell line. We show the gating strategy in Supplementary Figure 1. We sorted cells with a single-cell sort purity mask directly in Matrigel-coated 96-well plates containing 200 μl of media (as indicated later). For single-cell cloning, we cell sorted one cell per well (Figure 1).
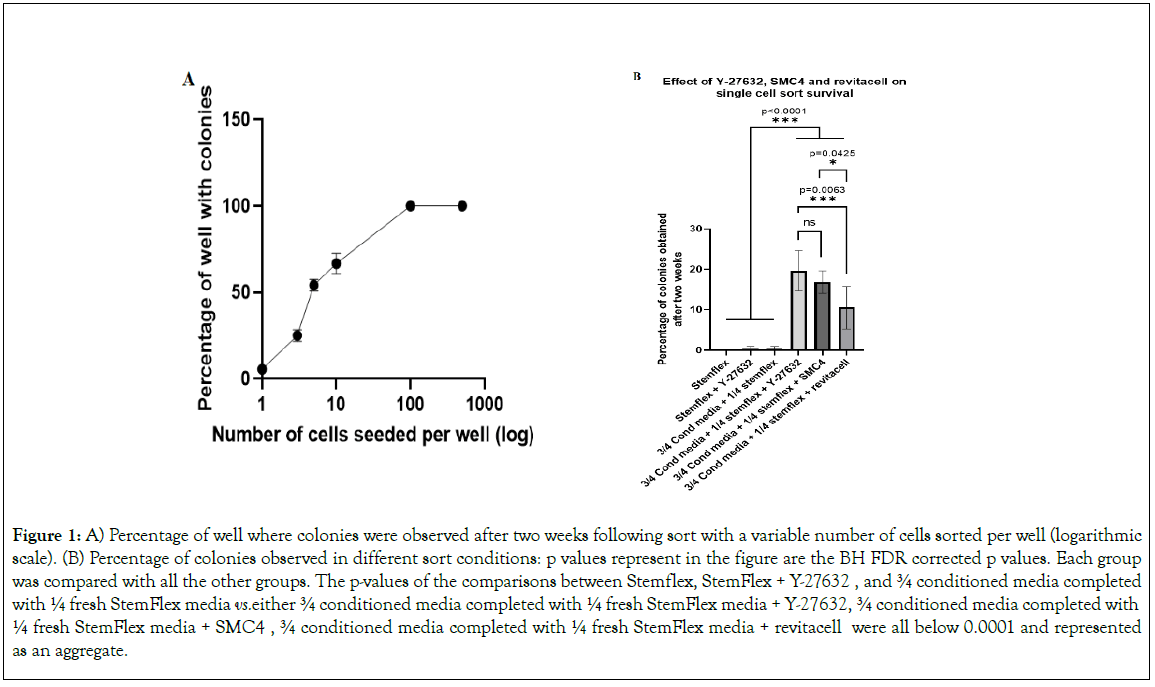
Figure 1: A) Percentage of well where colonies were observed after two weeks following sort with a variable number of cells sorted per well (logarithmic scale). (B) Percentage of colonies observed in different sort conditions: p values represent in the figure are the BH FDR corrected p values. Each group was compared with all the other groups. The p-values of the comparisons between Stemflex, StemFlex + Y-27632 , and ¾ conditioned media completed with ¼ fresh StemFlex media vs. either ¾ conditioned media completed with ¼ fresh StemFlex media + Y-27632, ¾ conditioned media completed with ¼ fresh StemFlex media + SMC4 , ¾ conditioned media completed with ¼ fresh StemFlex media + revitacell were all below 0.0001 and represented as an aggregate. Half of the media was changed 48 h post sort in all conditions and the following two days. Complete media was changed every day after.
Media conditioning and MVs isolation
Based on the increased stability of β-FGF in the StemFlex medium (up to 72 h according to the manufacturer), we produced a “conditioned” StemFlex medium by culturing HE iPSC Line at 40%-50% confluence for 24 h to avoid the complete depletion of media components. This conditioned medium was centrifuged at 300 g for 10 min, followed by a 20 min centrifugation at 2000 g to eliminate cells and cell debris [18]. After these two centrifugations, the conditioned medium was stored for up to 2 weeks at 4°C and labeled as “conditioned medium”. We isolated MVs from the conditioned medium by ultracentrifugation for 90 min at 100.000 g using UltraClear Tubes (#344058, Beckman Coulter) and an SW32Ti rotor k factor-204 (Beckman Coulter) in an Optima XPN-80 ultracentrifuge. The supernatant of ultracentrifugation was saved and labeled as MV-“depleted media”. We resuspended the pelleted MVs in a minimal medium volume (up to 1.5 ml for 300 ml of conditioned media) and stored medium and MVs for up to 2 weeks at 4°C prior to use. We recorded the volume of the original conditioned (spent) medium and diluted purified MV in a volume of fresh StemFlex medium equivalent to the volume of the original spent culture medium.
Qualitative and quantitative flow cytometry analysis of MVs
All media fractions and MVs prepared as described previously were analyzed using a Cytek Aurora spectral flow cytometer set up to optimally detect Apogee calibration beads (ApogeeMix#1493, Apogee). A double threshold was applied on Side Scatter Height (SSC-H) and fluorescence B2-H corresponding to 528/10 emission from blue laser (488 nm) excitation (B2 channel).
IPS validation by scorecard
We investigated the stemness and pluripotency of iPS post-sort using the TaqMan hPSC Scorecard Panel (Life Technologies) [19]. Briefly, undifferentiated cells were grown as described above and processed according to the manufacturer’s manual and published literature. For random differentiation into the three primary tissue lineages, we generated embryoid bodies (EBs) by 15 days of culture in low adherence 6-well plates in DMEM/F12 (Gibco,#10565-018) complemented with 20% KnockOut serum replacement (Gibco,#10828-010), MEM non-essential amino acids (Gibco,#11140-050) (1 mM) and β-mercaptoethanol (Gibco,#21985-023). We extracted total RNA from undifferentiated and differentiated cells using the TRIzol Reagent and RNeasy MinElute kit (Qiagen). We used DNAse I (Thermofisher) to remove contaminating DNA and used 1μg of purified RNA for reverse transcription using a high-capacity cDNA reverse transcription kit (Thermofisher). We performed Quantitative PCR on a 7500 fast Real-Time PCR system from Applied Biosystems.
We analyzed data online on the proprietary software of Life Technologies that calculates the relative score for iPSCs based on comparing expression profiles to a reference standard [20].
Immunofluorescence and confocal microscopy imaging
We confirmed the pluripotency of undifferentiated cells using the Pluripotent Stem Cell 4-Marker Immunocytochemistry Kit (Thermofisher,#A24881) according to the manufacturer’s recommendation. Briefly, cells were cultured on chamber slides (NuncTM Lab-TekTM Chamber Slide System, Thermofisher) coated with Matrigel and were fixed and permeabilized each for 15 minutes using the kit-provided buffers. Slides were then blocked with the kit blocking solution for 30 minutes before incubation with primary antibody for 3 h at 4°C. After rinsing with wash buffer, we incubated cells with secondary antibody for 1 h at room temperature and rinsed with the wash solution. We counterstained and mounted cells using Vectashield Antifade Mounting Medium with DAPI.
We stained differentiated cells with Human Three Germ Layer 3-Color Immunocytochemistry Kit (R&D Systems) according to the manufacturer’s recommendation. Briefly, we differentiated hIPSCs cultured in chamber slides into each of the three germ layers according to the protocol of the Human Pluripotent Stem Cell Functional Identification Kit (R&D Systems®, Catalog#SC027B) [21]. We stained ectoderm differentiated cells with Northern LightsTM (NL) 557-conjugated Otx-2 and NL493-conjugated SOX1. We stained mesoderm differentiated cells with NL557-conjugated Brachyury and NL637-conjugated HAND1. We stained endoderm differentiated cells with NL637-conjugated SOX17 and NL493-conjugated GATA-4. Counterstain was done using VECTASHIELD Antifade Mounting Medium with DAPI (Dako).
We analyzed immunostainings on Zeiss LSM 780 confocal microscope (Carl Zeiss) using ZEN Black software for image acquisition and analysis. We acquired all images with a pinhole set at 1 airy unit (AU) with a digital gain of 0. Specific setup of the confocal microscope is given for each picture in Supplementary Table 1. We acquired pictures of undifferentiated cells with a 63X oil immersion objective and pictures of differentiated cells with a 40X oil immersion objective.
We analyzed colonies with an EVOS FL auto microscope (Thermofisher) using 4X and 10X objectives and phase-contrast with a 10X objective.
Genomic stability analysis
Genomic DNA was isolated using Qiagen DNeasy kit (Qiagen,#69506) according to the manufacturer’s recommendation and quantified on Nanodrop. 290 ng of each sample DNA was used as a template to run the qPCR test on 7500 fast Real-Time PCR system from Applied Biosystems according to the manufacturer’s recommendation against a control DNA provided in the hPSC Genetic Analysis Kit (StemCell Technologies,#07550).
IPS surface marker analysis using LEGENDScreen
BioLegend’s LEGENDScreenTM Human PE Kit, which contains 361 PE-conjugated lyophilized monoclonal antibodies specific for human cell surface markers and their ten matched isotype controls, was used according to manufacturer recommendation on the HE iPSC line. Briefly, after antibodies reconstitution with water, cells were detached with ReLesR (Stemcell technologies) for 5 min as described above for single-cell sort. Cells were finally resuspended in the staining solution and filtered through a 40 μm cell strainer. 3.105 cells were added to each well of the plates containing the reconstituted antibodies and stained for 30min at 4°C. Cells were washed twice with staining buffer, centrifuged at 500 g for 10min, and rapidly flicked to remove liquid. Cells were then fixed using the provided fixation buffer for 10min and washed two additional times with staining buffer (Thermofisher). We acquired 100 μl per well of stained cells with an Acea Novocyte flow cytometer. We estimated the percentage of cells stained for a given marker by comparing the fluorescence intensity of the antibody-stained cells with that of matched isotype control. We analyzed data with Novoexpress from Acea Bioscience (Supplementary Table 2).
Statistical analysis
We calculated the correlation between the number of cells seeded per well and colonies formation at 6 days post-sorting by linear regression. We calculated P with a two-tailed test from the correlation coefficient score using all data. In order to compare differences among groups we fitted a general ANOVA model adding contracts between each group in comparison, and subsequently corrected the significance level for multiple tests using Benjamini and Hochberg (BH) False Discovery Rate (FDR).
Results
Cell isolation seems to impair more single cell survival than sort stress
We adjusted several parameters to reduce the stress induced by cell sorting. First, we reduced the number of surface markers analyzed to minimize any effects, such as receptor binding-induced activation. To this end, we sought to define a minimal set of markers to define IPSc.
When analyzed at day 21 and 35 post reprogramming, we could detect 40% of TRA1-60- SSEA4+ cells, 2,1% of cells were Tra-1- 60+ SSEA4+ and 24,5% of Tra-1-60+ SSEA4-. We performed further staining’s and sorts by gating only on cells expressing Tra- 1-60, which we use as a marker of pluripotent cells. We limited the lengths of the cell staining and sorting procedures to minimize their impact on stem cells. We established on HE iPSC line that for TRA-1-60 and SSEA4, 15-minute staining was as good as 1-hour staining and performed all staining with an incubation time of 15 min (Supplementary Figure 1c).
We investigated how cell sorting affected the survival of cells sorted from the well-established IPS line HE iPSC or the ESC lines ES4 and H1 (Figure 1). For each cell line, we sorted 500, 100, and 10 cells per well into 8 wells, and 5, 3, and 1 cell per well in 24 wells in a 96-well plate containing 200 μl of fresh media complemented with Y-27632 (Figure 1a). Two weeks post-sort, the wells wermnj examined, and wells displaying cell colonies were enumerated. Wells seeded with 500 or 100 cells per well systematically succeeded at growing colonies in each well (8 out of 8 wells displayed colonies for each cell line). Wells seeded with 10, 5, 3 or 1 cell, had significant decreased cell colonies counts at each stepdown in number of seeded cells (66.66+/-5.8%, 54.16+/- 3.4%, 25+/-3.4% and 5.55+/-1.96% respectively). We assessed the regression between the number of wells two weeks post-sort and the number of cells used to seed the wells between 10 and 1 cell per well (we excluded 100 and 500 because they support 100% cell growth) and found a linear regression (R=0.8439, P=2.43.10-5). These results indicate that, in our conditions, the colony-forming ability of sorted cells is around 5.5%.
Test of different sort-media to increase survival
Experiments described below were performed on well-established cell lines and hiPSCs lines derived from reprogrammed PBMCs 35 days post-transduction. We produced a conditioned StemFlex medium by culturing HE iPSC line at 40%-50% confluence for 24 h to avoid the depletion of media components. To ascertain the presence of all components and sufficient βFGF, we used ¾ conditioned media complemented with ¼ of “fresh” media. Data illustrated in Figure 1b and Supplementary Table 3 represent results obtained for HE iPSC and two newly reprogrammed hiPSCs lines (total three cell lines). We performed all sorts in a 96-well plate with 95 wells seeded with only one cell and one reference-well seeded with 100 cells. The analysis was done only in the 95 wells seeded with one cell. Conditioned media alone was insufficient to improve cell survival and colony formation. We, therefore, tested the use of Y-27632, SMC4 cocktail, and Revitacell to complement the conditioned media. There were no statistical differences in survival and colony formation for cells cultured with the three supplements despite slightly better results using Y-27632 (p=0.0945 between Y-27632 and Revitacell) in our conditions (Figure 2).
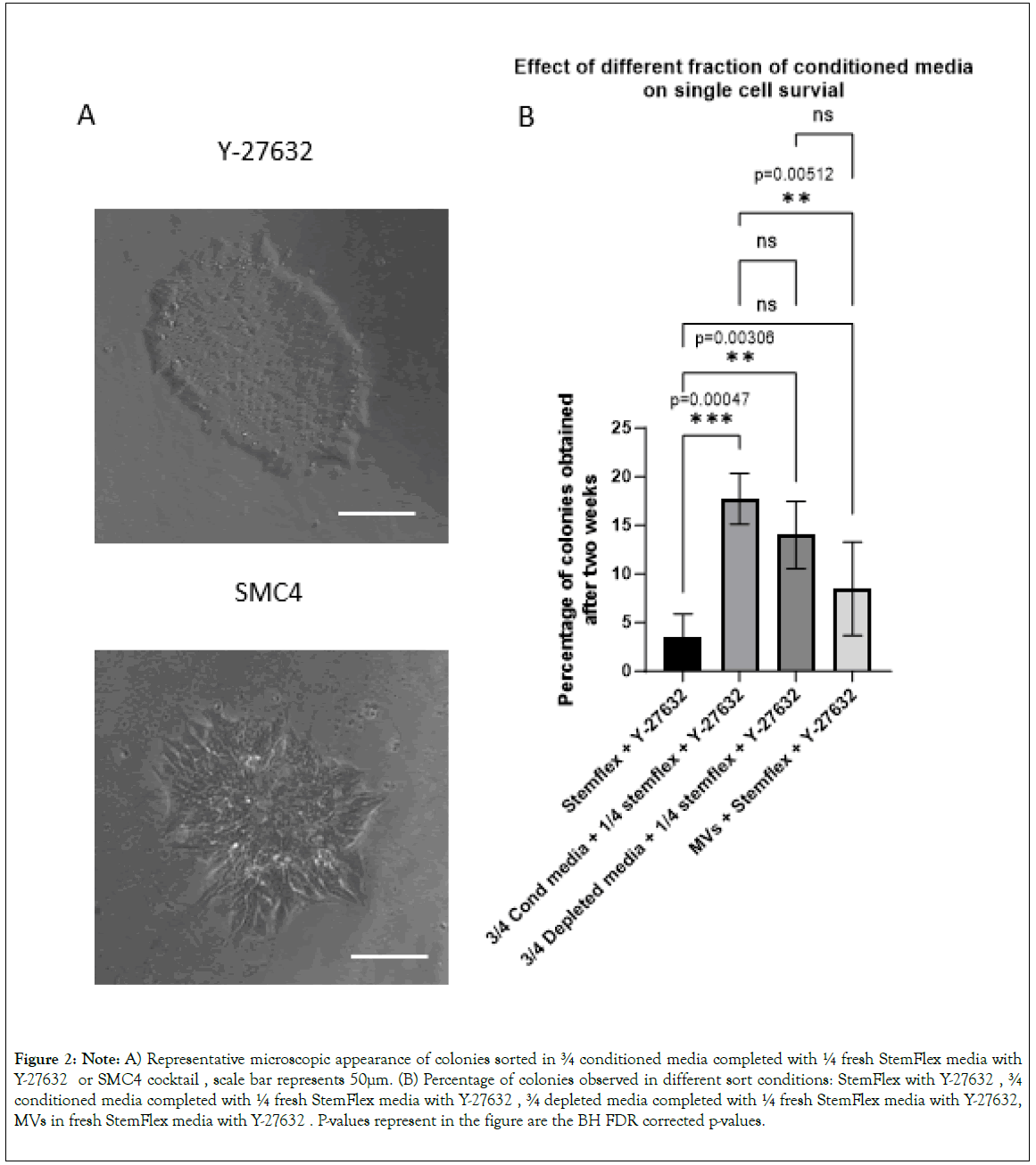
Figure 2: Note: A) Representative microscopic appearance of colonies sorted in ¾ conditioned media completed with ¼ fresh StemFlex media with Y-27632 or SMC4 cocktail, scale bar represents 50μm. (B) Percentage of colonies observed in different sort conditions: StemFlex with Y-27632, ¾ conditioned media completed with ¼ fresh StemFlex media with Y-27632, ¾ depleted media completed with ¼ fresh StemFlex media with Y-27632, MVs in fresh StemFlex media with Y-27632. P-values represent in the figure are the BH FDR corrected p-values.
The microscopic analysis of the colonies obtained after six days in wells seeded with a single cell clearly showed a better morphologic aspect of the cells and colonies when we cultured sorted cells in the presence of Y-27632 compared to the SMC4 cocktail (Figure 2A). In our setup, culturing single-cell sorted hiPSCs was most efficient using ¾ of conditioned media complemented with ¼ of fresh StemFlex media with Y-27632. We then investigated further the role of the conditioned medium.
Fraction of “conditioned media” leading to cell survival
Considering that hPSCs release a significant amount of MVs and that MVs are involved in many aspects of cell-to-cell communication, we investigated if secreted MVs were responsible for the survival of single cells seeded in a conditioned medium. Therefore, to isolate the effect of MVs, we generated a fresh culture medium complemented with MVs isolated from conditioned media and depleted MVs from conditioned media to isolate the effect of other soluble factors.
First, we analyzed the MVs released by the PSCs using the Cytek Aurora flow cytometer. We set up the cytometer using ApogeeMix reference beads, comprising silica beads of 180 nm, 240 nm, 300 nm, 590 nm, 880 nm, and 1300 nm in diameter, and latex green fluorescent beads of 110 nm and 500 nm diameter (Supplementary Figure 2a). The threshold cutoff of analysis was defined using 40 nm filtered water. Unlike PBS, which contained few detectable particles, (81+/-8 events/μl, n=3), StemFlex contained more detectable vesicles (18077+/-1410 events/μl, n=3) and conditioned media contained the highest amount of detectable vesicles (65694+/-10914 events/μl, n=9) reflecting the vesicles production by cells in culture. Ultracentrifugation of the conditioned medium depleted 91% of the microparticles, reducing their number to 5449+/-1339 events/μl (n=6). Pelleted MVs, when reconstituted to their original volume, reached a concentration of 74450+/-8610 events/μl (n=8), a number not significantly different from their original concentration (p=0,0006) (Supplementary Figure 2).
We tested the effect of different media fractions on the formation of colonies from single-cell after cell sorting. We performed this analysis on three “newly” reprogrammed cell lines 35 days post-transduction and on HE iPSC line (4 cell lines total). We performed all sorts in a 96-well plate with 95 wells seeded with only 1 cell-per well and 1 well seeded with 100 cells as reference. Conditioned media+Y-27632 and depleted media+Y-27632 gave the highest number of well containing cell colonies 18.68+/- 2.27% and 14.73+/- 3%, respectively compared to StemFlex+Y-27632 (3.68+/-2.06% (p=0.0002 and p=0.0025). The number of colony- positive wells was not different in cells cultured in MVs-containing media+Y-27632 (8.94+/-4.15%) compared with cells cultured StemFlex+Y-27632 (P=0.111) (Figure 2B and Supplementary Table 4). Altogether, those data indicate that the survival observed when using conditioned media is mostly due to hPSCs-released soluble factors that remain in the MVs-depleted fraction and are not associated with MVs.
Stemness validation and genome stability
We ascertained the stemness of derived hiPSCs clones by performing several validation assays on colonies obtained in each sort condition (Figure 3). First, we used the TaqMan hPSC scorecard assay to verify that cells did express pluripotency RNAs and could differentiate in all three germ layers (Figure 3a, Supplementary Figure 3a and 3b). In all conditions, we validated at least one clone by scorecards. We validated three different clones cultured in conditioned media, one from an established cell line and two from newly derived hiPSC. For MVs-depleted conditioned media, we validated one clone derived from an established hiPSC line and one from a freshly derived hiPSC. For supplemented culture medium, we validated one clone from a newly derived hiPSC. All clones were validated using scorecards. We further characterized two conditioned-media-derived clones by immunocytofluorescence staining (1 established cell line and one newly derived cell line).
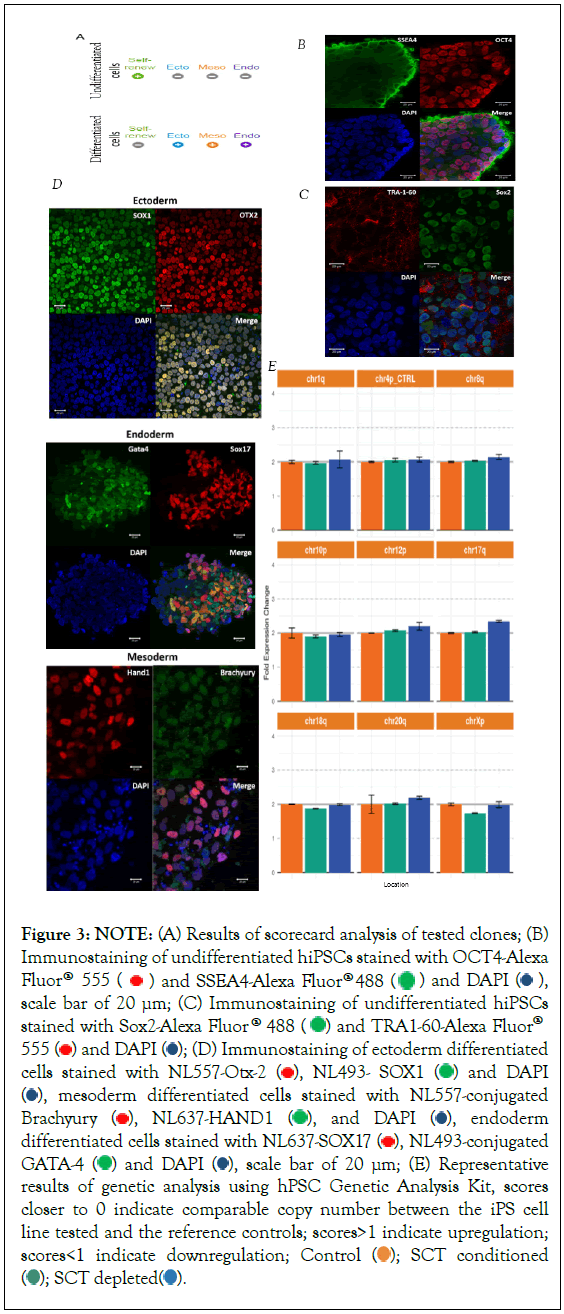
Figure 3: NOTE: (A) Results of scorecard analysis of tested clones; (B) Immunostaining of undifferentiated hiPSCs stained with OCT4-Alexa Fluor® 555  and SSEA4-Alexa Fluor® 488
and SSEA4-Alexa Fluor® 488  and DAPI
and DAPI  scale bar of 20 μm; (C) Immunostaining of undifferentiated hiPSCs stained with Sox2-Alexa Fluor® 488
scale bar of 20 μm; (C) Immunostaining of undifferentiated hiPSCs stained with Sox2-Alexa Fluor® 488  and TRA1-60-Alexa Fluor® 555
and TRA1-60-Alexa Fluor® 555  and DAPI
and DAPI  (D) Immunostaining of ectoderm differentiated cells stained with NL557-Otx-2
(D) Immunostaining of ectoderm differentiated cells stained with NL557-Otx-2  , NL493- SOX1
, NL493- SOX1  and DAPI
and DAPI  , mesoderm differentiated cells stained with NL557-conjugated Brachyury
, mesoderm differentiated cells stained with NL557-conjugated Brachyury  , NL637-HAND1
, NL637-HAND1  , and DAPI
, and DAPI  , endoderm differentiated cells stained with NL637-SOX17
, endoderm differentiated cells stained with NL637-SOX17  , NL493-conjugated GATA-4
, NL493-conjugated GATA-4  and DAPI
and DAPI  , scale bar of 20 μm; (E) Representative results of genetic analysis using hPSC Genetic Analysis Kit, scores
closer to 0 indicate comparable copy number between the iPS cell line tested and the reference controls; scores>1 indicate upregulation; scores<1 indicate downregulation; Control
, scale bar of 20 μm; (E) Representative results of genetic analysis using hPSC Genetic Analysis Kit, scores
closer to 0 indicate comparable copy number between the iPS cell line tested and the reference controls; scores>1 indicate upregulation; scores<1 indicate downregulation; Control  SCT conditioned
SCT conditioned  .
.
As shown in Figure 3b, undifferentiated cells stained positively for Oct4, Sox2, SSEA4, and Tra1-60. Their differentiated counterparts were stained positively for Otx2 and Sox1, Brachyury and Hand1 and Sox17 and Gata-4 for ectoderm, mesoderm, and endoderm differentiated cells, respectively (Figure 3c).
We tested the stability of the genome of sorted cells using a qPCR that detects the eight most common karyotypic abnormalities reported in hPSCs. Our results showed genome integrity in all clones tested in all sort conditions (Figure 3d).
Together those results show that in our setup, single-cell sorting allows the cloning of cells that can form colonies of pluripotent stem cells with no noticeable genome alteration.
Identification of receptors present on hPSCs potentially responsible for improved survival
We analyzed many surface markers expressed on hPSCs using the LEGENDScreen assay (Supplementary Table 2). We intended to identify a specific cell-surface receptor that could be responsive to soluble factors released by hPSCs and could affect cell survival. Several markers analyzed in the LegendScreen play a role in cell survival, such as Cytokine-Cytokine receptor interaction and PI3K- Akt signaling.
For instance, we could identify 9 receptors expressed (in more than 20% of the cells) on the surface of the HE iPSC line that are involved in the PI3K-akt pathway (CD29 (99,49%), CD49f (98.05% of cells), CD49e (69,14%), EphA2 (52,86%), CD49b (38,49%), CD49c (31,38%), CD49a (25,59%), CD221 (46,91%) and CD51(71,47%) (Supplementary Table 2). 46.91% of HE iPSC cells) expressed the insulin-like receptor 1 (IGFR1b, CD221 and 71.47% of cells expressed CD51.
Discussion
Manual passaging techniques for Stem Cell colonies were significantly improved by switching from traditional trypsin or collagenase to new enzymatic digestion methods with dispase and accutase [22,23]. More recently, non-enzymatic, EDTA-based methods or proprietary methods have improved cell detachment and passaging [24]. These new approaches have significantly improved cell survival following cell digestion required for cell passage. Manual selection of clones is highly subjective and prone to maintaining the heterogeneity of colonies found in the small aggregates that can contain un-reprogrammed or partially reprogrammed cells. This time-consuming method is hardly applicable to deriving many clones needed to screen genetically modified cells. In that regard, single-cell sorting is the only method that allows isolating “true” stem cell clones. One of the significant challenges remaining for applying this method to hPSC is the poor survival and colony formation of single-sorted cells. Previous publications have reported improved methods for single-cell sorting of cells culture on feeder layers of mouse embryonic fibroblasts (MEFs) or improved conditions for well-established cell lines [25-27]. Recent method employed microfluidic instruments for efficient and reliable survival, but do not apply with conventional flow cytometry [28]. Here we report a protocol that improves cell survival and can be applied to well-established cell lines and newly reprogrammed hiPSCs.
HPSCs are particularly sensitive to culture conditions [29]. Therefore, we decided to minimize the time between the cell’s detachment and the re-seeding of sorted cells. As maximum staining was obtained with an antibody incubation time of only 15 minutes at 37°C, we could minimize the time between detachment and re-seeding to 30 minutes. The Tra-1-60/ SSEA4 staining of reprogrammed cells conforms to reported results [30]. Because Tra- 1-60 expression has been reported to be the best marker of cells that will undergo entire reprogramming, cells were cloned solely based on their expression of this marker [31].
Under these conditions, we sorted a various number of cells per well, and in agreement with previous reports, demonstrated that when multiple cells are sorted together, cell survival and colony growth are greatly improved [4]. This demonstrates that the single- cell-dissociation and sort-stress are not solely responsible for increased death of hiPSCs post-sort, but physical isolation plays an essential role. One possible explanation for the improved survival in cell-sorted as multiple cells that we referred to as “crowd effect” was previously reported as a ‘neighbor model’ and “social behavior”, where “hPSCs survive better with neighboring cells than in isolation” [3,25]. These studies found that single-cell- cloning efficiency can be significantly improved when MEFs are present [25]. They refer to the work of Phadnis et al. to hypothesize that these effects may stem from signals that may include the direct sensing of neighboring cells or via cytokines/chemokines release [3].
We, therefore, used “conditioned media” (complemented with fresh media at a ratio of 3 to 1 to ensure the availability of fresh nutrients) to improve the cell survival post-sort. We demonstrate that the addition of conditioned media alone was insufficient to ensure cell survival. We, therefore, tested the addition of previously reported molecules that improve the survival of hiPSCs isolated as single cells [7]. Unlike previously published, in our conditions, Y-27632 and SMC4 cocktail showed better results than Revitacell, but Y-27632 treatment gave rise to colonies which, upon optical microscopy evaluation, showed a morphology typical of healthy and undifferentiated hPSCs colonies.
We demonstrated that in the presence of ROCK inhibitors (Y- 27632), single-cell-sorted hPSCs had a higher survival rate in StemFlex medium conditioned with HE iPSC line rather than with fresh medium. This technique was used to derive clones from well-established cell lines and from “newly” reprogrammed hiPSCs. This provides significant advantages both for facilitating and streamlining the hiPSCs derivation process and clonal isolation of genetically engineered cells compared to the manual isolation process. Despite the significance of our results, our tests were performed on a limited number of cell lines, and those results should be validated on a larger number of cell lines. Though it is noteworthy to state that while we employed Matrigel in the experiment shown in this manuscript, we successfully employed this method with Geltrex (data not shown) which constitutes a more reliable animal free substitute. These finding are consistent with previous report on the topic [27]. Also, in the recent past numerous commercial solutions were developed to improve single stem cell survival and should be compared to our method.
To understand the mechanism underlying this “Crowd effect” and the reason for this improved survival, we tested which elements in the “conditioned media” enhance the survival. hPSCs release MVs in vivo and in vitro. We isolated MVs from the conditioned media by ultracentrifugation for 90 min at 100.000 g. We then tested independently the fraction “depleted” of MVs and the purified MVs (complemented with fresh media) separately [11,12]. Our data demonstrate that most survival effect observed is conserved by the “depleted” fraction, hinting that soluble factors are primarily responsible for the effect (soluble proteins).
By screening for the cell surfaceome of hPSCs using the LEGENDScreen assay, we tried to identify receptors that conditioned media could activate. Our surface marker analysis using LEGENDScreen agrees with previously reported studies, and its results are typical of that of primed pluripotent state [32,33].
The activation of the PI3K Akt pathway is essential for the survival of pluripotent stem cells, and we found the expression of 9 receptors expressed at the surface of our hPSCs involved in this pathway [34]. Receptor CD49f, for example, is known to bind to LAMA, LAMB, and LAMC and stimulate the expansion of stem cells [35]. EphA2 was found expressed on our cells, and Ephrin proteins and receptors have also been shown to stimulate the growth of Stem cells [36]. Multiple cytokines such as FGF19, SDF-1, and CFS-1 were previously reported as expressed by hPSCs and implicated in cell survival and even essential to mobilize hPSCs and maintain their stemness [37-39]. We, therefore, expected to find the corresponding receptors expressed on our cells. Surprisingly, our surface marker analysis showed the expression of CD184 (CXCR4) (receptor of SDF-1α) in only 2.39% of cells and CXCR7 (alternative receptor of SDF-1α) in only 1,38% of cells. These surprising results were compatible with previously published data [33]. It is noteworthy that the team of M. Roederer showed that flow cytometry chemokine receptor staining on lymphocytes is strongly affected by the isolation method and best achieved when staining is performed at 37°C and not 4°C as recommended by Biolegend for LEGENDScreen [40]. Though using LEGENDScreen, other groups reported a stronger expression of CXCR4 (38%) on embryonic stem cell lines [32]. Other investigations showed that hiPSC lines display little if any expression of CXCR4 or CXCR7, while embryonic stem cell lines tend to have higher expression. Those receptors were reported to have an important role for differentiation when overexpressed, and therefore a restricted expression is more in line with pluripotency [41]. Other markers like CD51 (ITVGA) and IGFR1 were also found to be expressed on the surface of hPSCs. All these receptors were shown to be implicated in self-renewal and pluripotency maintenance [42]. In consequence, as we identified multiple potential receptors that could be responsible for stem cell survival and stemness maintenance, we hypothesize that the effect observed with conditioned media does not rely on a single pathway or cytokine-cytokine receptor interaction but, as previously proposed on a finely balanced combination of multiple factors and signals secreted by the HE iPSC line and their respective receptors [38].
Conclusion
In summary, this research challenges the notion that cell-sorting stress is the primary factor leading to decreased survival or genetic instability in Human Pluripotent Stem Cells (hPSCs) post-sort. This stress can be reduced by adjusting parameters during cell sorting, including minimizing the number of surface markers analyzed and optimizing staining procedures, to mitigate stress. Notably, we introduced a “crowd effect” by sorting multiple cells together, demonstrating a significant improvement in cell survival and colony growth, countering the belief that single-cell sorting stress is the main cause of reduced survival. The investigation of conditioned media supplemented with various molecules revealed that Y-27632 significantly enhanced survival and morphological characteristics of sorted cells. Furthermore, the study delves into the role of Microvesicles (MVs) in cell survival, suggesting that soluble factors within the MVs-depleted fraction contribute significantly to the observed effects. The identification of multiple receptors associated with the PI3K-Akt pathway and other signaling pathways further supports the hypothesis that the enhanced survival is a result of a balanced combination of signals from various factors secreted by hPSCs. Overall, the findings present a streamlined protocol for improving cell survival during single-cell sorting, offering affordable and valuable insights for advancing stem cell research and applications.
Authors Contributions
Christophe Raynaud contributed to the Conceptualization, Methodology, Validation, Formal analysis, Investigation, Resources, Writing manuscript, Supervision. Sharefa Al-mannai contributed to the Investigation.
Authors Disclosure
Authors have no conflict of interest to report.
Funding
This research was funded by Sidra research department funds.
References
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861-872.
[Crossref] [Google Scholar] [PubMed]
- Chin AC, Padmanabhan J, Oh SK, Choo AB. Defined and serum-free media support undifferentiated human embryonic stem cell growth. Stem Cells Dev. 2010;19(6):753-761.
[Crossref] [Google Scholar] [PubMed]
- Phadnis SM, Loewke NO, Dimov IK, Pai S, Amwake CE, Solgaard O, et al. Dynamic and social behaviors of human pluripotent stem cells. Sci Rep. 2015;5:14209.
[Crossref] [Google Scholar] [PubMed]
- Chen Y-H, Pruett-Miller SM. Improving single-cell cloning workflow for gene editing in human pluripotent stem cells. Stem Cell Res. 2018;31:186-192.
[Crossref] [Google Scholar] [PubMed]
- Chen KG, Mallon BS, McKay RDG, Robey PG. Human pluripotent stem cell culture: considerations for maintenance, expansion, and therapeutics. Cell Stem Cell. 2014;14(1):13-26
[Crossref] [Google Scholar] [PubMed]
- Krawetz R, Taiani JT, Liu S, Meng G, Li X, Kallos MS, et al. Large-scale expansion of pluripotent human embryonic stem cells in stirred-suspension bioreactors. Tissue Eng Part C Methods. 2010;16(4):573-582.
[Crossref] [Google Scholar] [PubMed]
- Valamehr B, Abujarour R, Robinson M, Le T, Robbins D, Shoemaker D, et al. A novel platform to enable the high-throughput derivation and characterization of feeder-free human iPSCs. Sci Rep. 2012;2:213.
[Crossref] [Google Scholar] [PubMed]
- Howden SE, Thomson JA, Little MH. Simultaneous reprogramming and gene editing of human fibroblasts. Nat Protoc. 2018;13(5):875-898.
[Crossref] [Google Scholar] [PubMed]
- Steiner D, Khaner H, Cohen M, Even-Ram S, Gil Y, Itsykson P, et al. Derivation, propagation and controlled differentiation of human embryonic stem cells in suspension. Nat Biotechnol. 2010;28(4):361-364.
[Crossref] [Google Scholar] [PubMed]
- Tang SW, Thomas A, Murai J, Trepel JB, Bates SE, Rajapakse VN, et al. Overcoming Resistance to DNA-Targeted Agents by Epigenetic Activation of Schlafen 11 (SLFN11) Expression with Class I Histone Deacetylase Inhibitors. Clin Cancer Res. 2018;24(8):1944-53.
[Crossref] [Google Scholar] [PubMed]
- Zhou J, Ghoroghi S, Benito-Martin A, Wu H, Unachukwu UJ, Einbond LS, et al. Characterization of Induced Pluripotent Stem Cell Microvesicle Genesis, Morphology and Pluripotent Content. Sci Rep. 2016;6:19743.
[Crossref] [Google Scholar] [PubMed]
- Desrochers LM, Bordeleau F, Reinhart-King CA, Cerione RA, Antonyak MA. Microvesicles provide a mechanism for intercellular communication by embryonic stem cells during embryo implantation. Nat Commun. 2016;7:11958.
[Crossref] [Google Scholar] [PubMed]
- Hewitt Z, Forsyth NR, Waterfall M, Wojtacha D, Thomson AJ, McWhir J. Fluorescence-activated single cell sorting of human embryonic stem cells. Cloning Stem Cells. 2006;8(3):225-34.
[Crossref] [Google Scholar] [PubMed]
- Grievink HW, Luisman T, Kluft C, Moerland M, Malone KE. Comparison of Three Isolation techniques for human peripheral blood mononuclear cells: Cell recovery and viability, population composition, and cell functionality. Biopreserv Biobank. 2016;14(5):410-415.
[Crossref] [Google Scholar] [PubMed]
- Tanabe K, Nakamura M, Narita M, Takahashi K, Yamanaka S. Maturation, not initiation, is the major roadblock during reprogramming toward pluripotency from human fibroblasts. Proc Natl Acad Sci USA. 2013;110(30):12172-12179.
[Crossref] [Google Scholar] [PubMed]
- Stoyanova E, Mourdjeva M, Kyurkchiev S. Early selection of human fibroblast-derived induced pluripotent stem cells. Biotech Biotech Equi. 2015;29(5):942-948.
- Pomeroy JE, Hough SR, Davidson KC, Quaas AM, Rees JA, Pera MF. Stem cell surface marker expression defines late stages of reprogramming to pluripotency in human fibroblasts. Stem Cells Transl Med. 2016;5(7):870-882.
[Crossref] [Google Scholar] [PubMed]
- Morales-Kastresana A, Telford B, Musich TA, McKinnon K, Clayborne C, Braig Z, et al. Labeling extracellular vesicles for nanoscale flow cytometry. Sci Rep. 2017;7(1):1878.
[Crossref] [Google Scholar] [PubMed]
- Ferrer M, Corneo B, Davis J, Wan Q, Miyagishima KJ, King R, et al. A multiplex high-throughput gene expression assay to simultaneously detect disease and functional markers in induced pluripotent stem cell-derived retinal pigment epithelium. Stem Cells Transl Med.
[Crossref] [Google Scholar] [PubMed]
- Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144(3):439-452.
[Crossref] [Google Scholar] [PubMed]
- Gagne AL, Maguire JA, Gandre-Babbe S, Chou ST, Tasian SK, Loh ML, et al. Generation of a human Juvenile myelomonocytic leukemia iPSC line, CHOPi001-A, with a mutation in CBL. Stem Cell Res. 2018;31:157-160.
[Crossref] [Google Scholar] [PubMed]
- Draper JS, Smith K, Gokhale P, Moore HD, Maltby E, Johnson J, et al. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat Biotechnol. 2004;22(1):53-54.
[Crossref] [Google Scholar] [PubMed]
- Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 2007;25(6):681-686.
[Crossref] [Google Scholar] [PubMed]
- Beers J, Gulbranson DR, George N, Siniscalchi LI, Jones J, Thomson JA, et al. Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat Proto. 2012;7(11):2029-2040.
[Crossref] [Google Scholar] [PubMed]
- Singh AM. An Efficient Protocol for Single-Cell Cloning Human Pluripotent Stem Cells. Front Cell Dev Biol. 2019;7:11.
[Crossref] [Google Scholar] [PubMed]
- Nicholas CR, Gaur M, Wang S, Pera RA, Leavitt AD. A method for single-cell sorting and expansion of genetically modified human embryonic stem cells. Stem Cells Dev. 2007;16(1):109-117.
[Crossref] [Google Scholar] [PubMed]
- Chen YH, Pruett-Miller SM. Improving single-cell cloning workflow for gene editing in human pluripotent stem cells. Stem cell res. 2018;31:186-192.
[Crossref] [Google Scholar] [PubMed]
- Tristan CA, Hong H, Jethmalani Y, Chen Y, Weber C, Chu PH, et al. Efficient and safe single-cell cloning of human pluripotent stem cells using the CEPT cocktail. Nat Protocl. 2023;18(1):58-80.
[Crossref] [Google Scholar] [PubMed]
- Konagaya S, Ando T, Yamauchi T, Suemori H, Iwata H. Long-term maintenance of human induced pluripotent stem cells by automated cell culture system. Sci Rep. 2015;5:16647.
[Crossref] [Google Scholar] [PubMed]
- Bharathan SP, Manian KV, Aalam SMM, Palani D, Deshpande PA, Pratheesh MD, et al. Systematic evaluation of markers used for the identification of human induced pluripotent stem cells. Biol Open. 2017;6(1):100-108.
[Crossref] [Google Scholar] [PubMed]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Sasaki A, Yamamoto M, et al. Induction of pluripotency in human somatic cells via a transient state resembling primitive streak-like mesendoderm. Nat Commun. 2014;5:3678.
[Crossref] [Google Scholar] [PubMed]
- Collier AJ, Panula SP, Schell JP, Chovanec P, Plaza Reyes A, Petropoulos S, et al. Comprehensive cell surface protein profiling identifies specific markers of human naive and primed pluripotent states. Cell Stem Cell. 2017;20(6):874-90.e7.
[Crossref] [Google Scholar] [PubMed]
- Adkar SS, Wu CL, Willard VP, Dicks A, Ettyreddy A, Steward N, et al. Step-wise chondrogenesis of human induced pluripotent stem cells and purification via a reporter allele generated by crispr-cas9 genome editing. Stem cells. 2019;37(1):65-76.
[Crossref] [Google Scholar] [PubMed]
- Hossini AM, Quast AS, Plotz M, Grauel K, Exner T, Kuchler J, et al. PI3K/AKT signaling pathway is essential for survival of induced pluripotent stem cells. PloS one. 2016;11(5):e0154770.
[Crossref] [Google Scholar] [PubMed]
- Krebsbach PH, Villa-Diaz LG. The role of integrin α6 (CD49f) in stem cells: More than a conserved biomarker. Stem Cells Dev. 2017;26(15):1090-1099.
[Crossref] [Google Scholar] [PubMed]
- Genander M, Frisen J. Ephrins and Eph receptors in stem cells and cancer. Curr Opin Cell Biol. 2010;22(5):611-616.
[Crossref] [Google Scholar] [PubMed]
- Krejci P, Kunova M, Kubikova I, Trantirek L, Kozubik A, Dvorak P. Expression of FGF19 in human embryonic stem cells. Stem cells. 2013;31(11):2582-2584.
[Crossref] [Google Scholar] [PubMed]
- Jiang Z, Li Y, Ji X, Tang Y, Yu H, Ding L, et al. Protein profiling identified key chemokines that regulate the maintenance of human pluripotent stem cells. Sci Rep. 2017;7(1):14510.
[Crossref] [Google Scholar] [PubMed]
- Hong D, Ding J, Li O, He Q, Ke M, Zhu M, et al. Human-induced pluripotent stem cell-derived macrophages and their immunological function in response to tuberculosis infection. Stem Cell Res Ther. 2018;9(1):49.
[Crossref] [Google Scholar] [PubMed]
- Berhanu D, Mortari F, De Rosa SC, Roederer M. Optimized lymphocyte isolation methods for analysis of chemokine receptor expression. J Immunol Methods. 2003;279(1-2):199-207.
[Crossref] [Google Scholar] [PubMed]
- Reid JC, Tanasijevic B, Golubeva D, Boyd AL, Porras DP, Collins TJ, et al. CXCL12/CXCR4 signaling enhances human PSC-derived hematopoietic progenitor function and overcomes early in vivo transplantation failure. Stem Cell Rep. 2018;10(5):1625-1641.
[Crossref] [Google Scholar] [PubMed]
- Teng C-F, Jeng L-B, Shyu W-C. Role of insulin-like growth factor 1 receptor signaling in stem cell stemness and therapeutic efficacy. Cell Transplant. 2018;27(9):1313-9.
[Crossref] [Google Scholar] [PubMed]
Citation: Raynaud CM, Al-Mannai S (2023) Use of Media Conditioning for Robust Single Cell Sorting of Induced Pluripotent Stem Cells. Single Cell Biol. 12:068.
Copyright: © 2023 Raynaud CM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Sources of funding : This research was funded by Sidra research department funds
