Indexed In
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- Ulrich's Periodicals Directory
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Proquest Summons
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2025) Volume 0, Issue 0
Structural Modeling Analysis and Functional Characteristics of Two F10 Mutations (Asp413Asn and Gly420Arg)
Osayd Zohud1, Siba Shanak2, Husam Sallam1, Zaidoun Salah1 and Hisham Darwish1,3*2Faculty of Science, Arab American University, Jenin, Palestine
3Department of Medical Laboratory Sciences, Faculty of Allied Medical Sciences, Arab American University, Jenin, Palestine
Received: 09-Apr-2025, Manuscript No. JBDT-25-28661; Editor assigned: 11-Apr-2025, Pre QC No. JBDT-25-28661 (PQ); Reviewed: 25-Apr-2025, QC No. JBDT-25-28661; Revised: 02-May-2025, Manuscript No. JBDT-25-28661 (R); Published: 09-May-2025, DOI: 10.4172/2155-9864.25.16.056
Abstract
Factor X (FX), a vitamin K-dependent serine protease, plays a crucial role in coagulation, and mutations in the F10 gene can lead to FX deficiency. This study aimed to investigate the functional and structural consequences of two missense mutations, c.1237 G>A (Asp413Asn) and c.1258 G>A (Gly420Arg). Wild-type and mutant FX constructs were transfected into HEK293 cells to evaluate intracellular protein levels, secretion rates, and FX activity, as well as mRNA expression. The Asp413Asn mutant exhibited significantly lower intracellular protein levels, while Gly420Arg levels were similar to wild type. However, Asp413Asn showed higher secretion compared to both Gly420Arg and wild type. Both mutants displayed significantly reduced FX activity compared to wild type. Bioinformatics analysis revealed that Asp413 and Gly420 are highly conserved in the catalytic domain, and molecular modeling indicated that both mutations lead to damaging structural alterations. These findings suggest that the distinct functional impairments associated with Asp413Asn and Gly420Arg mutations contribute to the pathology of FX deficiency, highlighting the importance of these residues in maintaining FX activity.
Keywords
FX deficiency; Molecular genetics; Bleeding disorders; Palestine; Modeling; F10 gene
Introduction
Factor X is a vitamin K-dependent clotting factor which serves as precursor for serine protease (FXa) and constitutes a major clotting component in the blood coagulation mechanism activated by intrinsic and extrinsic cascades in the common pathway of thrombin generation [1]. It forms an active complex with factor Va which catalyzes the production of thrombin from prothrombin, which converts fibrinogen into fibrin [2, 3]. It is exclusively synthesized by the liver and secreted in circulation as a pre-peptide following modification by a vitamin K-dependent enzyme system [4]. In circulation, proteolytic cleavage of a 40 a.a. activation peptide yields the active form of Factor X [4, 5].
The mature protein is composed of two peptides, light chain (17 kDa) and heavy chain (45 kDa) linked by disulfide bond [6]. The mature structure consists of several functional domains including a serine protease domain responsible for its catalytic activity [7].
Factor X deficiency is a rare disease inherited as an autosomal recessive genetic disorder with worldwide incidence of ~1 per million [4], however, in the Palestinian community, the incidence reaches about 20 per million [8]. The F10 gene is located on Chromosome 13q34c and consists of 8 exons, each encoding a specific domain of the protein [9]. Several mutations have been identified in the F10 gene, predominantly missense mutations [10]. These mutations define two types of FX deficiency: type I, where FX protein is not produced with no activity detected, and type II with FX protein expressed but with limited to no activity [11]. We previously reported a novel missense mutation in a Palestinian patient (Asp413Asn coding sequence numbering, Asp189 based on chymotrypsin numbering system) located in the catalytic domain of the protein [8].
Even though over 170 mutations have been previously reported in the F10 gene, limited knowledge is available concerning the structural and functional consequences of these mutations [12]. In the present study, we generated two recombinant FX missense mutant proteins including the Asp413Asn mutation and another previously reported mutation (Gly420Arg, Gly380 in mature FX and based on chymotrypsin numbering system) similarly located in the catalytic domain [8, 13], and studied their molecular structural and functional implications.
This work offers novel insights through focusing on the unique genetic variants in the F10 gene by applying an integrative approach combining molecular, biochemical, and computational analyses. This includes the use of molecular dynamics simulations and in silico bioinformatic tools to examine how these mutations impact the structural stability and function of the Factor X protein. Moreover, we compare the expression levels and functional activity of the two mutant proteins using cell-based assays, revealing new information about how these mutations affect protein secretion and coagulation activity. These insights provide extended understanding of the molecular pathology of Factor X deficiency and offer potential clinical implications for managing bleeding disorders in affected patients.
Materials and Methods
FX site-directed mutagenesis
The vector containing human full-length F10 cDNA (WT-F10.cDNA) was obtained from Origene (CAT.NO: RC208506). Site-directed mutagenesis was used to introduce the Asp413Asn and Gly420Arg mutations using Quick Change Lightning Site-Directed Mutagenesis Kit (Stratagene, LA Jolla, CA, USA) according to the manufacturer’s protocol. The sequences of the primers used to generate the mutations were as follows: Forward primer 5’-CCCTGGCAGGCATTCTCCTGCTTGGTG-3’ and reverse primer 5’-CACCAAGCAGGAGAATGCCTGCCAGGG-3’ for the Asp413Asn mutation, and forward primer 5’-AGGGGGACAGCAGGGGCCCGCAC-3’ and reverse primer 5’-GTGCGGGCCCTGCTGCTGTCCCCCT-3’ for the Gly420Arg mutation. The wild-type and mutant constructs were transformed into DH5α competent cells, and isolated plasmids were sequenced to confirm the intended nucleotide change.
Cell culture and transfection
Human Embryonic Kidney cells (HEK293) were cultured and maintained in Dulbecco Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum under 5% CO2. Three independent transient transfection experiments were done using ViaFect transfection reagent (Promega, Madison, WI, USA) as advised by the manufacturer for both mutants and the wild-type plasmid constructs, in addition to mock plates that served as negative control. Additionally, FX protein from a normal blood sample, quantified and validated as wild-type by ELISA, was used as a positive control for FX protein detection by Western blot. Transfected cells were incubated for 48 hours in serum-free DMEM media as described before [4]. At the end of the incubation period, a sample of transfected cells was treated with Cyclohexamide for 1 hour. The supernatant conditioning media was collected in pre-chilled tubes, centrifuged at 3000 g for 10 minutes at 4°C, and stored at -80°C. Transfected cells were lysed using RIPA lysis buffer supplemented with protease inhibitor cocktail and PMSF.
Real-time PCR
RNA was extracted from HEK293 transfected cells using TRI reagent (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s protocol. F10 cDNA was synthesized using 1 μg of total RNA using Maxima First Strand cDNA Synthesis Kit (Thermo Fisher Scientific Inc., Waltham, MA, USA). RT-qPCR was done to quantitate F10 RNA reflected by the synthesized cDNA from each sample using SYBR™ Green PCR Master Mix (Thermo Fisher Scientific Inc., Waltham, MA, USA). As an internal control, β-actin gene was used to normalize cDNA quantification results. Two sets of primer pairs were used in RT-qPCR to quantify the expression levels of F10 and β-actin genes. For F10, the primers were forward 5′-TGTCCAGCAGCTTCATCATC-3′ and reverse 5′-GCGGTGACCTTGGTGTAGAT-3′, whereas for β-actin, the forward primer was 5′-TTCCAGCCTTCCTTCCTGGG-3′ and the reverse primer was 5′-TTGCGCTCAGGAGGAGCAAT-3′.
Recombinant FX activity and expression assays
The activity of recombinant FX expressed by the various constructs was measured using PT clotting assay and adjusted based on the protein levels detected by ELISA. Conditioned media was concentrated 10-fold using a speed-vac system, and FX-deficient plasma was used for the clotting reaction. The plasma was mixed with concentrated conditioned media in a 1:1 ratio and PT was tested using HumaClot Jr blood coagulation machine. The protein expression level in the cell lysates by the recombinant FX constructs was detected by Western blot analysis. Briefly, samples were electrophoresed in 15% SDS-polyacrylamide gel, blotted on polyvinylidene membrane, and probed with primary monoclonal mouse anti-human FX antibody (Proteintech Cat#: 66753-1-Ig), followed by probing with a secondary goat anti-mouse globulin tagged with horseradish peroxidase (BETHYL). Finally, the signal was detected using Clarity Western ECL (enhanced luminol-based chemiluminescent) substrate reagent (Bio-Rad) and evaluated using a gel documentation system (Bio-Rad). The level of recombinant mutant FX protein secreted in the culture media and in the lysates of transfected cells was measured by ELISA using Human Factor X ELISA Kit (Abcam) and compared to the relevant protein levels of the wild-type construct. The relative variation in protein levels, RNA levels, and protein activity between the wild-type and the two mutants was assessed using Student's t-test.
Evolutionary conservation of Asp413 and Gly420 in FX protein
The conservation and putative functional impact of the two mutations of interest on the FX protein (Uniprot code P00742) were analyzed using online bioinformatic tools PolyPhen-2, SIFT, HOPE, and COBALT [14–17]. The numbers referenced in the text followed the chymotrypsin numbering system.
Molecular modeling
As structural reference for the simulations of the human factor X protein, we used the X-ray structure of the human des (1-45) factor Xa at 2.2 Å resolution (RCSB:1HCG; 7). The wild-type protein was mutated to the two mutant forms (Asp413Asn and Gly420Arg) using the Pymol software [18]. Through a pairwise alignment of the FASTA sequences for the experimentally retrieved sequence and the sequence found at the RCSB database, we found that Asp413 corresponds to Asp189, and Gly420 corresponds to Gly196 in the chain A of the crystallographic evidence.
Molecular Dynamics (MD) simulations for all systems were performed with the GROMACS 4.5.5 package [19, 20], using the AMBER99SB force field [21] and the TIP3P water model [22]. Each wild-type or mutant protein was placed in a cubic water box of 8.5 nm box dimensions with 0.10 mol/l NaCl added to neutralize the system. The total size of the simulated systems was around 13,200 atoms. The setup of this system was analogous to a previous study [23].
Periodic boundary conditions were employed. Columbic interactions were evaluated using a short-range cut-off of 10 Å, and long-range interactions were treated by the Particle Mesh Ewald (PME) summation method [24]. The non-bonded Lennard-Jones interactions were computed using a smooth cut-off of 10 Å. The integration time step was set to 2 fs. The temperature was kept at 310 K by applying leapfrog stochastic dynamics forces with a damping coefficient of 0.1 ps-1 [25].
At first, the simulated systems were energy-minimized for 50,000 steps using the steepest descent algorithm. The tolerance was set to 1.0 kJ mol-1 nm-1. After that, the system was heated to 310 K for 4 ps. Next, each system was subjected to 1 ns equilibration in the NVT ensemble with harmonic restraints applied to all protein atoms using a force constant of 1000 kJ mol-1 nm-2. With restraints kept, each system was further equilibrated for 500 ps in the NPT ensemble, and then for another 500 ps without restraints.
Plain MD simulations for each system were performed over 100 ns each, like in [23]. Via inspecting the RMSD values for the protein (including side chains) with respect to the reference structures in the three systems, the protein structures in the three simulated systems were well converged after 50 ns of MD simulation. Thus, 100 ns simulations were conducted where three structures were stable enough for further analysis.
Results
F10 mRNA expression
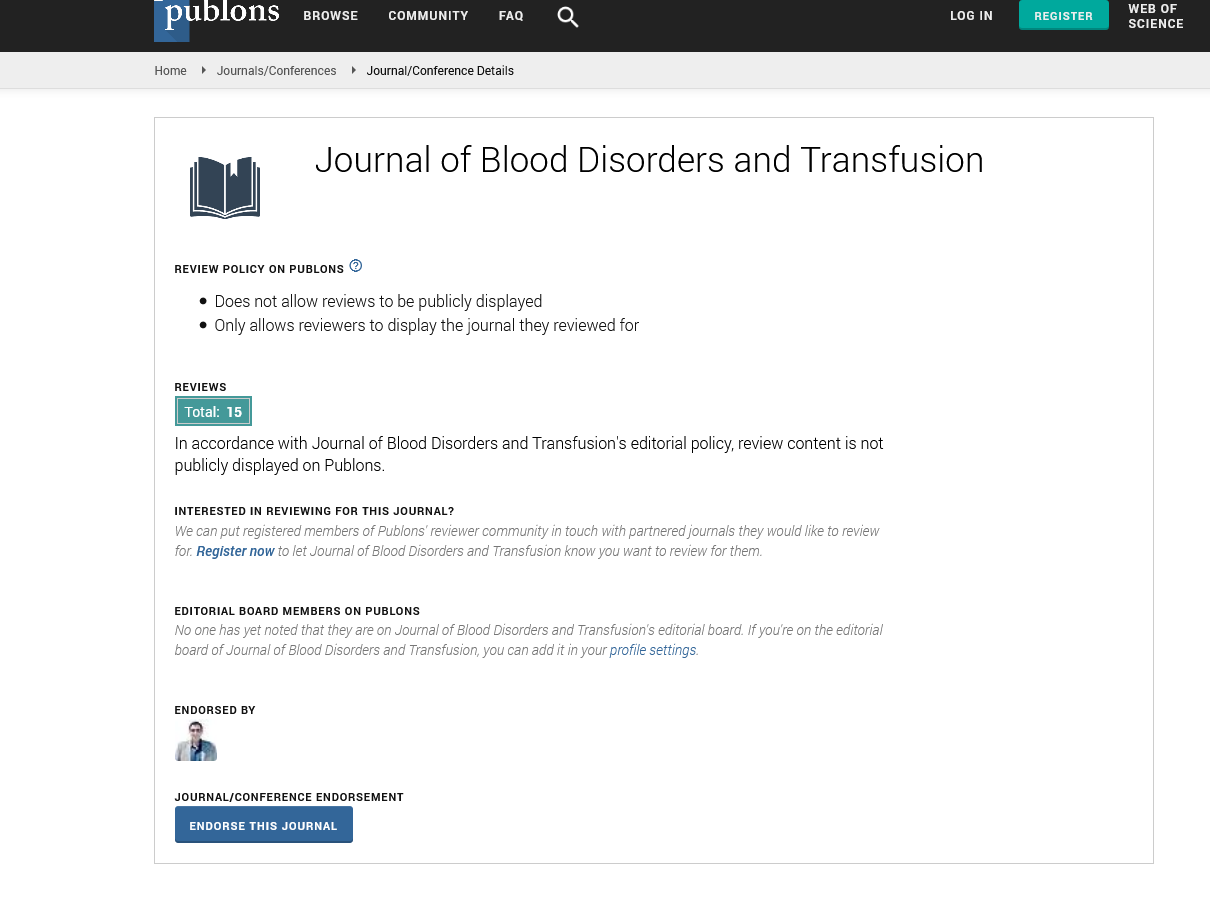
Since some mutations affect the expression of genes [26], we tested the effect of the two indicated mutations on F10 mRNA level. Total RNA was extracted from HEK293 cells transfected with WT, Gly420Arg and Asp413Asn constructs. RT-qPCR was used to quantify F10 mRNA expression levels from the three recombinant vectors normalized to the expression level of β-actin. The results from two independent replicates indicated no significant difference in F10 mRNA expression levels from cells transfected with the Asp413Asn (p=0.2) mutant variant while the Gly420Arg mutant showed lower RNA level compared to WT at (p=0.05) as shown in (Figure 1).
Figure 1: Expression of FX mutants. RT-qPCR analysis showing relative expression levels of FX mRNA in cells transfected with the three indicated recombinant constructs and mocked transfected cells. The three replicate values for each recombinant are shown with small circles and the star represents the mean value ± SD. The expression activity of the two mutant constructs were quantitated relative to the expression level in WT transfected cells.
Effect of Asp413Asn and Gly420Arg mutations on FX protein expression
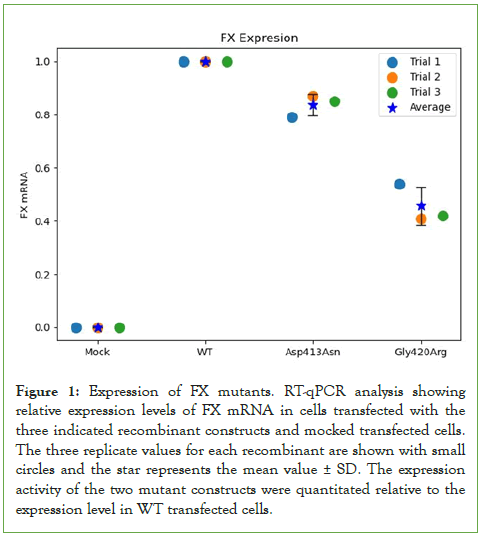
To investigate the effect of the indicated mutations on FX protein levels, equal amounts of protein from cell lysate transfected with the three constructs and mock-transfected cells were loaded on SDS-PAGE and processed with Western blotting analysis. FX protein was similarly detected as expected at ~50 kDa molecular weight from all cells transfected with the FXWT, FXAsp413Asn and FXGly420Arg constructs, with no FX protein detected in mock-transfected cells (Figure 2A). Evidently, no factor X protein bands were detected in transfected cells in the presence of cycloheximide indicating that the detected bands represent newly synthesized FX protein (Figure 2B). The detected protein bands represent the heavy chain of FX protein since the protein gel was run under reducing conditions in the presence of beta-mercaptoethanol. No FX protein band could be detected in the lane loaded with lysates from mock transfected cells. The expression of GAPDH protein (~37 kDa) in all cell lysates including mock-transfected cells was used as control as shown in (Figure 2C).
Figure 2: Protein expression of FX variants. A. Western blot showing the expression of FX protein (~50 kDa) in cell lysates of transfected cells with Mock, Gly420Arg (M2), Asp413Asp (M1) and Wild Type (WT) constructs, In addition to a Positive Control (PC). B. FX variants protein expression after Cyclohexa-mide treatment for 1 hour showing the absence of FX expression in the indicated transfected cells C. Expression of the GAPDH gene ( ) in the lysates Cyclohexamide treated (CHX+) and non-treated (CHX-) cells. The protein size marker in kDa is shown on the right side of the
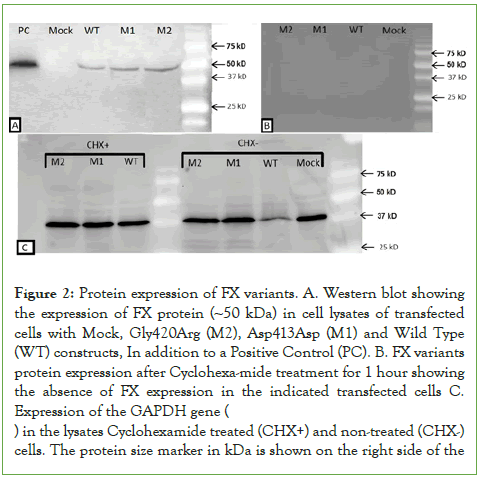
GAPDH expression seems similar in all transfected cells including mock-transfected cells in the presence or absence of cycloheximide. Factor X proteins were directly quantitated using ELISA, in cell lysates and conditioned culture media of cells transfected with either Asp413Asn, Gly420Arg mutants or wild-type constructs. Figure 3A shows the expression level of Gly420Arg was comparable to that of WT (44.6 ng/mL (108%) and 43.4 ng/mL (100%) respectively, p=0.2), whereas the Asp413Asn mutant expression was significantly lower compared to the WT (36.3 ng/mL (84%) p=0.001). This could be related to variation in the rate of translation of the Asp413Asn or stability of the mutant protein.
Figure 3: (A). FX protein levels in cell lysates. Quantitative evaluation of FX protein levels in cell lysates transfected with the indicated FX variants quantitated by ELISA assay. The values of three replicate measurements are shown with small circles and the star represents the mean value with SD. The protein levels of two mutant variants were calculated relative to the protein level of WT treated cells. (B). Secreted FX protein levels. Similar quantitative evaluation of secreted FX protein in the conditioned media of the same cells transfected with the indicated recombinant constructs. The FX protein levels from cells transfected with the two mutant constructs were calculated relative to the protein level from WT transfected cells.
Level and activity of secreted FX
To assess the level of secreted FX, we similarly evaluated the corresponding proteins level in the conditioned culture media of transfected cells with both mutants and the WT constructs using ELISA. The level of secreted WT protein was 0.93 ng/mL (set as 100%) compared to 1.32 ng/mL for the Asp413Asn mutant (142%, p=0.03) and 0.94 ng/mL for Gly420Arg mutant (101%, p=0.5), as shown in (Figure 3B). These results indicate the difference of FX level between Asp413Asn mutant and the WT shown in the cell lysates could be related to variation in the rate of synthesis or secretion of this mutant in the culture media compared to the WT and the Gly420Arg mutant.
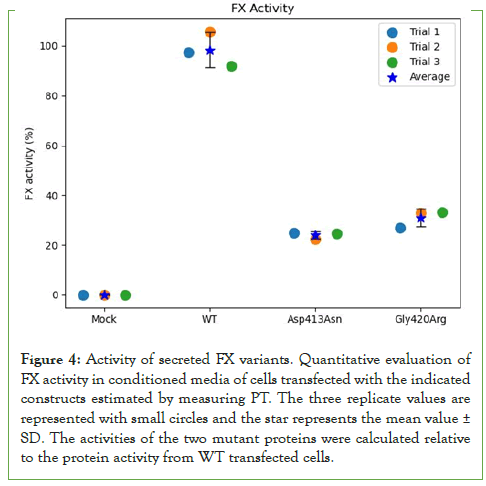
In addition, the effect of both mutations on FX clotting activity adjusted to FX antigen levels was directly evaluated compared to the WT. The Asp413Asn and Gly420Arg mutants showed 25.9% (PT=63 Sec) and 31.4% (PT=72 Sec) activity relative to WT set to 100% (PT=23 Sec) as shown in (Figure 4). These results show the mutant FX proteins had significantly lower activity than WT protein comparable to the activity levels measured in patients’ plasma.
Figure 4: Activity of secreted FX variants. Quantitative evaluation of FX activity in conditioned media of cells transfected with the indicated constructs estimated by measuring PT. The three replicate values are represented with small circles and the star represents the mean value± SD. The activities of the two mutant proteins were calculated relative to the protein activity from WT transfected cells.
Conservation and in silico analyses of Asp413 and Gly420
Conservation analysis of Asp413 and Gly420 in the F10 gene revealed that both amino acids are conserved in homologous species, suggesting that both residues play an important role in the structure and activity of the protein (Figure 5). Moreover, bioinformatics prediction tools (PolyPhen-2, PROVEAN, SIFT and HOPE), predicted both mutations to be damaging and likely leading to significant distortion of protein structure and function (data not shown).
Figure 5: Conservation analysis of (A) Asp413 and (B) Gly420 showing that both positions were conserved among various homologues species, the position of the discussed conserved amino acids is indicated by a black rectangles.
Structural analysis of the Asp413Asn and Gly420Arg FX mutants
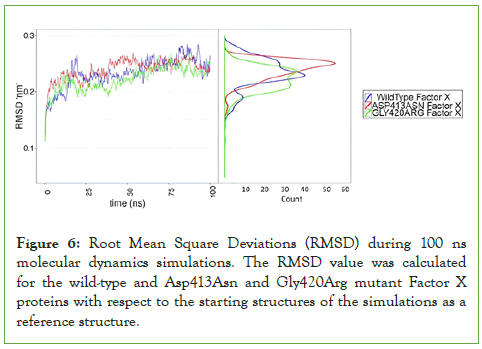
The three-dimensional structure modeling of both mutants was carried out in comparison to the wild type. The results showed similar overall structure compared to the wild type, while introducing structural variation within the catalytic site of the protein (data not shown). The wild-type and mutant proteins were all inspected for their structural stability and convergence for further downstream analysis via analyzing the RMSD values for the whole structures including side chains (Figure 6).
Figure 6: Root Mean Square Deviations (RMSD) during 100 ns molecular dynamics simulations. The RMSD value was calculated for the wild-type and Asp413Asn and Gly420Arg mutant Factor X proteins with respect to the starting structures of the simulations as a reference structure.
The RMSD values for the three structures were calculated in reference to the initial structures of the simulated proteins. The three structures converged well at 50 ns of simulation to ~0.25 nm from the reference structure, since they showed stably converged RMSD values at the indicated timeslot. The results indicate that the three systems are stable enough to be additionally analyzed for collective variables under question in this study. Since our above presented results show that Asp413Asn and Gly420Arg mutations have lower FX activity, we wanted to test whether this effect may be due to perturbations of the FX structure.
To this end, we inspected the Root Mean Square Fluctuation (RMSF) for the alpha carbons in the whole protein sequence during 100 ns long molecular dynamics simulations of the wild-type and mutant proteins (Snapshots were output each 1 ps; with a total of 100,000 simulated structures being analyzed).
Most of the results were comparable, except in some patches, where we noticed slightly increased RMSF values for some residues of the two mutants relative to wild type (Residues 123: 0.125 nm in Gly420Arg, 0.136 nm in Asp413Asn; compared to 0.081 nm in the wild type; residues 130-136: A mean of 0.172 ± 0.045 nm in Asp413Asn, compared to 0.068 ± 0.008 nm and 0.071 ± 0.014 nm, respectively, in wild type and Gly420Arg).
KS test was performed for the differences in RMSF values between the wildtype and any of the mutants, calculated for the alpha carbons in the stretch of residues 123 and 130-136. This helps to conclude the statistical significance for the differences in the stability between the wildtype and any of the two mutants by reading the RMSF values at each 1 ns interval (wildtype to Asp413Asn: D=0.20539, p-value <2.2e-16; wildtype to Gly420Arg: D=0.21324, p-value <2.2e-16). This suggests a less stable structure for the indicated residues in the mutants when compared to the wild type.
The aforementioned residues, with comparably higher RMSF values in the mutants than in the wildtype, occur in a single stretch of alpha-helix in the peptidase S1 domain, in close proximity to the catalytic triad of the same domain. The secondary structure was also inspected, using a trajectory of 250 snapshots throughout the 100 ns simulations. Results denote changes of the secondary structure between the wild type and either of the two mutants (Data not shown). The dynamic change in the secondary structure was not at the mutation interface, but rather at the surrounding protein stretches, where some drift in the secondary structure was noticed.
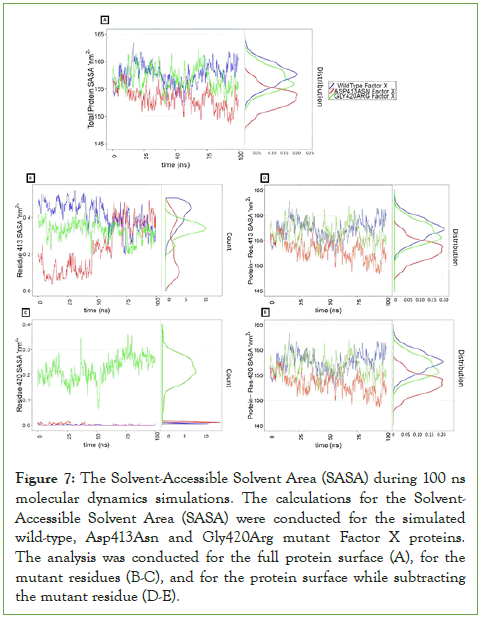
We performed Solvent Accessible Surface Area (SASA) calculations for the whole protein, for the protein while subtracting the SASA for the corresponding residue where the mutation took place, and for the residues where the mutation occurred. We noticed that the SASA value for the wild-type protein was relatively larger than the corresponding values for the two mutants (Figure 7), irrespective of what mutation took place and of whether the mutation induced a more hydrophilic residue (an average of ≈ 158 nm2 for the wild-type, compared to an average of ≈156 nm2 for the Gly420Arg mutant, and an average of ≈ 154 nm2 for the Asp413Asn mutant, see (Figure 7A)).
Subtracting the SASA for the target residues at the mutation interface did not affect the comparative differences between the wild-type and any of the two mutants see (Figure 7D-7E), which could be attributed to the very small surfaces of the different amino acids (ranging from 0-0.5 nm2, see (Figure 7B-7C)). It should be emphasized that the variations in the exposed surface areas in (Figure 7B-7C) barely account for fluctuations in the exposed solvent surface to one to few atoms of the amino acid. Overall, the noticeable difference in SASA between the whole protein surface of the wild-type and mutants indicates an overall change at the mutation interface that introduced a more hydrophobic environment upon mutation.
Figure 7: The Solvent-Accessible Solvent Area (SASA) during 100 ns molecular dynamics simulations. The calculations for the Solvent- Accessible Solvent Area (SASA) were conducted for the simulated wild-type, Asp413Asn and Gly420Arg mutant Factor X proteins. The analysis was conducted for the full protein surface (A), for the mutant residues (B-C), and for the protein surface while subtracting the mutant residue (D-E).
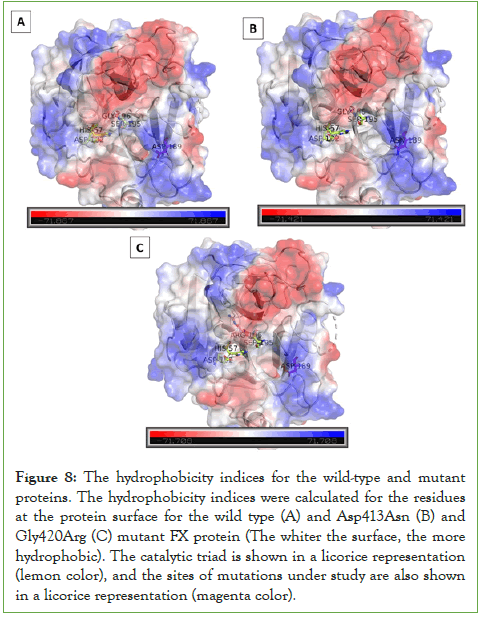
To further visually inspect the effect the mutations induced on the solvent accessible area, refer to (Figure 8). It compares the hydrophobicity indices for the residues at the protein surface (the whiter the patch, the more hydrophobic). Asp413Asn mutant (Figure 8B), where Asn189 refers to the respective mutated Asn413 reside and Gly420Arg mutant (Figure 8C), where Arg196 refers to the respective mutated Asn413 reside induced an increase in the hydrophobicity, when compared to the wild type; seen in whiter patches at the protein interface. This supports the results obtained from the SASA calculations. To further understand the impact of the reduced solvation upon mutation, hydrogen bonding status, where the mutation occurred, was also examined before and after the mutation. The side-chain N atoms of the mutant Asp413Asn induced an increase in the hydrogen bonding to the surrounding amino acid residues when compared to the WT Asp413. This was also the case upon mutating the Gly420 residue, where the mutant Asn420 showed an increased hydrogen bonding with the surrounding amino acids in comparison to the wild type. Still, H-bonds between the wild-type residue and the surrounding residues were found to be a bit stronger. The distribution of H-bond distances showed that the mean distance for H-bonds for the target residue to the surrounding ones was slightly shorter (≈0.275 nm) than the two mutants in residue 413 (≈0.288 nm). In residue 420, the H-bond distribution showed a higher peak height for the average H-bond distance in the wild type than in the Gly420Arg mutant. However, the wild type and the two mutants had a similar average (≈0.295 nm) in the H-bonding between the target residue and the surrounding residues in the protein.3.1. Subsection
Figure 8: The hydrophobicity indices for the wild-type and mutant proteins. The hydrophobicity indices were calculated for the residues at the protein surface for the wild type (A) and Asp413Asn (B) and Gly420Arg (C) mutant FX protein (The whiter the surface, the more hydrophobic). The catalytic triad is shown in a licorice representation (lemon color), and the sites of mutations under study are also shown in a licorice representation (magenta color).
Discussion
In the present study, we investigated the functional and structural consequence of two specific missense mutations in the factor X gene, one mutation caused by c.1237 G>A transition in exon 8 leading to Asp413Asn and the second mutation caused by similar c.1258 G>A transition leading to Gly420Arg with severe loss of FX activity [4]. Both mutations are in close proximity of the catalytic domain of FX protein and were predicted to have major impact on the structure and function of FX protein due to the nature of the amino acid shift from negatively charged to neutral side chain in one mutant and from small neutral to a bulky positively side chain in the other [6]. In addition, conservation analysis of both Asp413 and Gly420 among multiple species revealed that these amino acids are highly conserved in the FX protein and other serine proteases. This strongly suggests that both amino acids in addition to their relevant locations are crucial for FX structure and activity similar to other missense mutations in the gene [27, 28]. This prediction was highly supported by bioinformatics prediction tools (PolyPhen-2, PROVEAN, SIFT and HOPE) indicating that these mutations are likely damaging and may lead to significant distortion of protein structure and function. Clearly, mutations of highly conserved amino acids residues or a stretch of a conserved region is predicted to cause deficiency in protein activity as shown in mutations in the conserved loop 40 of several related proteins including Prothrombin, factor VII and factor IX [29–31]. A recombinant expression system was used to investigate the molecular mechanism of FX insufficiency due to the indicated mutations. The results showed no significant difference in F10 mRNA of Asp413Asn compared to the wild type with lower level in the case of the Gly420Arg mutant variant. These results indicate mild impact of the mutants on gene transcription. Similar findings were reported in previous studies that examined the impact of other missense mutations on F10 expression. One study showed the mRNA level of the Ala275Val FX mutant was 1.8-fold higher compared to the wild type [4], however, 5 other mutations in the gene showed similar intracellular level of F10 mRNA and protein production including Gly154Arg, Gly263Val, Val424Phe, Gly406Ser, Val424Phe, Val236Met, Arg387Cys and Gly406Ser compared to the wild type indicating no effect on gene expression and protein stability [6]. The Asp413Asn mutant protein was detected in cell lysate with significant reduction whereas the Gly420Arg mutant showed no significant difference in FX protein level compared to the wild type. This variation could be due to difference in stability between the two mutant proteins compared to the wild type or impairment of intracellular trafficking, similar to the Ala275Val mutant [4]. The level of secreted proteins from the two indicated mutants in this study showed variation compared to the wild type (Asp413Asn higher than the WT) similar to other mutant proteins seems to be influenced by the mutation type and location as shown in previous studies [28]. In addition, variation in the protein level secretion could be influenced by translation efficiency due to various gene mutations [32]. Direct functional testing (PT) revealed significant reduction in the Asp413Asn mutant protein coagulant activity (~25% compared with the wild type). Similarly, the FX activity of the Gly420Arg mutant was 31% compared to the wild type. These results indicate that both mutant proteins were successfully synthesized and released but their activities were drastically reduced. The significant decrease in both mutant proteins activity seems to be related to their inability to bind their substrate in the catalytic region since both mutations lie in close proximity to the catalytic triad (his 57, asp 102, ser 195; see (Figure 8)), including the Ala275Val FX mutant [4] and the A191V mutation in FVII gene, as suggested before [33–35]. Indeed, binding of FX to either pro-thrombin or FVa proteins occurs via cleaving some of their arginine residues. This indicates the Gly420Arg mutation induces a more positive interface that is less compatible in cleaving the target proteins [36–38]. Furthermore, the Asp413Asn mutation lies at the positively charged interface of the protein and might cause lower level of integrity in the protein structure, where negatively and positively charged stretches of the protein do not interact properly due to the compromised positive charge. In comparison, the level of secreted mutant FX proteins in the culture media was found to vary significantly due to other reported missense mutations which seem to have varying impacts on the trafficking and stability of the secreted proteins [4, 6]. Evidently, regardless of the variation of secreted FX mutant proteins, their reduced functional activity significantly exceeds the observed difference in the variation of their secretion efficiency [6].
In addition to examining the three-dimensional structure of the mutants in comparison to the wild type which showed some structural rearrangement in this region of the protein which houses the catalytic domain similar to the Ala275Val [4] and other mutants [6]. We inspected the Root Mean Square Fluctuation (RMSF) for the whole protein sequence in the wild type and mutants. The slight increase in the RMSF values for the two mutants when compared to the wild type suggests a less stable structure for the indicated residues in the mutants when compared to the wild type. On the other hand, a dynamic change in the secondary structure found in comparing the wild type with the two mutants was not at the mutation interface but rather at the surrounding protein stretches. This indicated a structural rearrangement seen also at the level of solvent accessible surface area and individual hydrogen bonding patterns that might impact the protein’s function.
Conclusion
In conclusion, two recombinant F10 mutant constructs, Gly420Arg and Asp413Asn with mutations located in the catalytic domain were generated to investigate their molecular structural and functional implications on the protein. The data revealed similar mRNA levels between both mutants compared to the wild type with an apparent insignificant lower level in case of the Gly420Arg mutant variant. However, significant difference in the expressed and secreted protein levels between the three indicated constructs was observed. Direct functional activity showed significant reduction of both Asp413Asn (~25%) and Gly420Arg (31%) mutants compared to the wild type. Structural analysis showed slight increase in the RMSF values for both mutants compared to the wild suggesting less stable structure for the indicated residues in the mutants compared to the wild type. Dynamic change in the secondary structure indicated structural rearrangement that might impact the protein’s function. Solvent Accessible Surface Area (SASA) values indicated introduction of a more hydrophobic environment associated with the mutation with changing hydrogen bonding. However, the average H-bond length was shorter for the wild type with the surrounding residues and with water, suggesting more stable hydrogen bonding in the wild type.
Author Contributions
Osayd Zohud was directly involved in performing all the biological lab work of this project (including relevant construct generation, transfection, RNA quantification and protein studies) and the relevant data analysis as part of his master’s project at AAUP. He also participated in constructing the first draft of the manuscript. Siba Shanak supervised all the protein structural and modeling work and drafted relevant sections of this part in the manuscript. Husam Salam provided technical support and expertise in the lab work. Zaidoun Salah provided assistance in performing major part of the biological data using his lab facility and took active role in reviewing and modifying the manuscript. Hisham Darwish served as the supervisor and coordinator of the whole project.
References
- Chafa O, Tagzirt M, Tapon-Bretaudière J, Reghis A, Fischer AM, LeBonniec BF. Characterization of a homozygous Gly11Val mutation in the Gla domain of coagulation factor X. Thrombosis research. 2009;124(1):144-148.
- Shinohara K, Adachi M, Matsui K, Matsuda K, Nagaya S, Morishita E. A case of factor X (FX) deficiency due to novel mutation V196M, FX Hofu. Int J Hematol. 2008;87:256-259.
- Cooper DN, Millar DS, Wacey A, Pemberton S, Tuddenham EG. Inherited factor X deficiency: molecular genetics and pathophysiology. Thromb Haemost. 1997;78(07):161-172.
- Sun N, Chen Y, Peng H, Luo Y, Zhang G. A novel Ala275Val mutation in factor X gene influences its structural compatibility and impairs intracellular trafficking and coagulant activity. Thromb Res. 2016;138:108-113.
- Messier TL, Pittman DD, Long GL, Kaufman RJ, Church WR. Cloning and expression in COS-1 cells of a full-length cDNA encoding human coagulation factor X. Gene. 1991;99(2):291-294.
- Nagaya S, Akiyama M, Murakami M, Sekiya A, Asakura H, Morishita E. Congenital coagulation factor X deficiency: Genetic analysis of five patients and functional characterization of mutant factor X proteins. Haemophilia. 2018;24(5):774-785.
- Padmanabhan K, Padmanabhan KP, Tulinsky A, Park CH, Bode W, Huber R, et al. Structure of human des (1-45) factor Xa at 2· 2 Å resolution. J Mol Biol. 1993;232(3):947-966.
- Smoom R, Abushkedim I, Darwish H. Identification of two Novel Mutations in the Factor X gene; A 5'Donor Splice-Site Mutation (IVS1+ 1G? T) and a missense mutation (Asp413Asn G> T) in unrelated palestinian factor X deficient patients. J Blood Disord Transfus. 2015;6:4.
- Furie B, Furie BC. The molecular basis of blood coagulation. Cell. 1988;53(4):505-518.
- Arita K, Niimi H, Yamagishi N, Ueno T, Kitajima I, Sugiyama T. Factor X heterozygous mutation in a patient with potential risk of bleeding: A case report. Medicine (Baltimore). 2018;97(23):e10950.
- Chatterjee T, Philip J, Nair V, Mallhi RS, Sharma H, Ganguly P, et al. Inherited factor X (Stuart–Prower factor) deficiency and its management. Med J Armed Forces India. 2015;71:S184-S186.
- Stenson PD, Mort M, Ball EV, Chapman M, Evans K, Azevedo L, et al. The Human Gene Mutation Database (HGMD®): Optimizing its use in a clinical diagnostic or research setting. Hum Genet. 2020;139:1197-1207.
- Vianello F, Lombardi AM, Boldrin C, Luni S, Girolami A. A new factor X defect (Factor X Padua 3): A compound heterozygous between true deficiency (Gly380→ Arg) and an abnormality (Ser334→ Pro). Thromb Res. 2001;104(4):257-264.
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248-249.
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012.
- Venselaar H, Te Beek TA, Kuipers RK, Hekkelman ML, Vriend G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics. 2010;11:1-10.
- Papadopoulos JS, Agarwala R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23(9):1073-1079.
- DeLano WL. The PyMOL molecular graphics system, version 1.8. Schrödinger, LLC. 2020.
- Hess B, Kutzner C, Van Der Spoel D, Gromacs EL. 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435-447.
- Lemkul JA. Introductory tutorials for simulating protein dynamics with GROMACS. J Phys Chem B. 2024;128(39):9418-9435.
- Lindorffâ?Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, et al. Improved sideâ?chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78(8):1950-1958.
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926-935.
- Schenkelberger M, Shanak S, Finkler M, Worst EG, Noireaux V, Helms V, et al. Expression regulation by a methyl-CpG binding domain in an E. coli based, cell-free TX-TL system. Phys Biol. 2017;14(2):026002.
- Darden T, York D, Pedersen L. Particle mesh Ewald: An N log (N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089-.
- Van Gunsteren WF, Berendsen HJ. A leap-frog algorithm for stochastic dynamics. Molecular Simulation. 1988;1(3):173-185.
- Lutz S, Brion C, Kliebhan M, Albert FW. DNA variants affecting the expression of numerous genes in trans have diverse mechanisms of action and evolutionary histories. PLoS Genet. 2019;15(11):e1008375.
- Jin Y, Cheng X, Zheng J, Xia H, Yang L, Wang M. A novel factor X mutation Cys81 by Arg and a reported factor VII polymorphism Arg353 replaced by Gln co-occured in a patient. Blood Coagul Fibrinolysis. 2018;29(1):67-74.
- Chikasawa Y, Shinozawa K, Amano K, Ogata K, Hagiwara T, Suzuki T, et al. Factor X M402T: A homozygous missense mutation identified as the cause of cross-reacting material-reduced deficiency. Int J Hematol. 2014;100:345-352.
- Giannelli F, Green PM, Sommer SS, Poon MC, Ludwig M, Schwaab R, et al. Haemophilia B: Database of point mutations and short additions and deletions—eighth edition. Nucleic Acids Res. 1998;26(1):265-268.
- Millar DS, Elliston L, Deex P, Krawczak M, Wacey AI, Reynaud J, et al. Molecular analysis of the genotype-phenotype relationship in factor X deficiency. Hum Genet. 2000;106:249-257.
- Akhavan S, Mannucci PM, Lak M, Mancuso G, Mazzucconi MG, Rocino A, et al. Identification and three-dimensional structural analysis of nine novel mutations in patients with prothrombin deficiency. Thromb Haemost. 2000;84(12):989-997.
- Davyt M, Bharti N, Ignatova Z. Effect of mRNA/tRNA mutations on translation speed: Implications for human diseases. J Biol Chem. 2023;299(9).
- Borensztajn K, Chafa O, Le Bonniec B, Wajcman H, Reghis A, Fischer AM, et al. Inherited factor VII deficiency: Identification of two novel mutations (A191V and T239P) in the catalytic domain. Thromb Res. 2005;116(2):115-120.
- Miyata T, Kojima T, Suzuki K, Umeyama H, Yamazaki T, Kamiya T, et al. Factor X Nagoya 1 and Nagoya 2: A CRM–factor X deficiency and a dysfunctional CRM+ factor X deficiency characterized by substitution of Arg306 by Cys and of Gly366 by Ser, respectively. Thromb Haemost. 1998;79(03):486-490.
- Isshiki I, Favier R, Moriki T, Uchida T, Ishihara H, Van Dreden P, et al. Genetic analysis of hereditary factor X deficiency in a French patient of Sri Lankan ancestry: in vitro: expression study identified Gly366Ser substitution as the molecular basis of the dysfunctional factor X. Blood Coagul Fibrinolysis. 2005;16(1):9-16.
- Dahlbäck B. Novel insights into the regulation of coagulation by factor V isoforms, tissue factor pathway inhibitorα, and protein S. J Thromb Haemost. 2017;15(7):1241-1250.
- Schuijt TJ, Bakhtiari K, Daffre S, DePonte K, Wielders SJ, Marquart JA, et al. Factor Xa activation of factor V is of paramount importance in initiating the coagulation system: lessons from a tick salivary protein. Circulation. 2013;128(3):254-266.
- Jackson CM. Structure and function of factor X: Properties, activation, and activity in prothrombinase. A retrospective in a historical context. J Thromb Thrombolysis. 2021;52(2):371-378.
Citation: Darwish H, Sallam H, Zohud O, Salah Z, Shanak S (2025). Structural Modeling Analysis and Functional Characteristics of Two F10 Mutations (Asp413Asn and Gly420Arg). J Blood Disord Transfus. 16:056.
Copyright: © 2025 Darwish H, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.