Indexed In
- Academic Journals Database
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- China National Knowledge Infrastructure (CNKI)
- Scimago
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- University Grants Commission
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research - (2020) Volume 12, Issue 2
rpoB, katG and inhA Genes: The Mutations Associated with Resistance to Rifampicin and Isoniazid in Egyptian Mycobacterium tuberculosis Clinical Isolates
Amal M Hosny1*, Hala M Abu Shady2 and Ayman K El Essawy32Microbiology Department, Faculty of Science, Ain Shams University, El-Khalyfa El-Mamoun Street, El-Abaseya, Egypt
3Microbiology Department, Ain Shams University Specialized Hospital (ASUSH), Ain Shams University, Cairo, Egypt
Received: 27-Jan-2020 Published: 09-Mar-2020, DOI: 10.35248/1948-5948.20.12.428
Abstract
In response to the huge number of people who die yearly due tuberculosis and the emergence of multidrug resistant (MDR) M. tuberculosis, accurate and rapid detection of this resistance can improve the situation. Relapsed patients in the current work represented significant percentages among rifampicin and isoniazid resistant isolates compared to other risk factors. Two molecular techniques (Genotype MTBDRplus assay and specific gene sequencing were used to detect associated mutations in TB drug resistant isolates. The genotypic profile of Multi-drug resistant (MDR) isolates showed missing of katG wild type 1 (WT1) band. Eighty percent of isoniazid mono-resistant isolates, showed katG MUT1, 20% showed katG MUT1 and inhA MUT1, 20% showed only inhA MUT1. The molecular techniques partly predicted the level of antibiotic resistance associated with katG and/or inhA gene mutations (for isoniazid) and rpoB gene mutation (for rifampicin). MTBDRplus could clearly detect rifampicin resistance among 66.7% of MDR isolates that showed mutation band rpoB MUT3 while 33.3% of them were considered as unknown, while 100% of mono-isoniazid resistant strains were detected. A mono-resistant rifampicin isolate did not show rifampicin mutation bands by Genotype MTBDRplus assay, but it showed unexpected mutation in codon 531 of rpoB by DNA sequence analysis, it can be considered as heteroresistant strain. Gene sequencing could detect resistance associated mutations mainly in codon 315 (katG gene), position -15 (inhA gene) for isoniazid resistance and codon 531 (rpoB gene) for rifampicin resistance.
Keywords
rpoB, katG, inhA, Rifampicin, Isoniazid, Genotype MTBDRplus assay
Introduction
Tuberculosis led to an estimated 1.2 million deaths among HIVnegative people and an additional 251000 deaths among HIVpositive people in the year 2018. As first line treatment, rifampicin and isoniazid are the key drugs in regimens used worldwide for the treatment of tuberculosis (TB). Drug-resistant TB continues to be a public health threat. The three countries with the largest share of the global burden were India (27%), China (14%) and the Russian Federation (9%). Globally, 3.4% of new TB cases and 18% of previously treated cases had multidrug resistant TB or rifampicinresistant TB (MDR/RR-TB), with the highest proportions (>50% in previously treated cases) in countries of the former Soviet Union [1].
Additionally, Due to the widespread use of anti-TB drugs, not only multidrug-resistant tuberculosis (MDR-TB) was spread but even extensively drug-resistant tuberculosis (XDR-TB) are now emerging and represents a considerable challenge to current TB prevention and control programs [2,3].
Moreover, the number of inaccurate diagnoses and inappropriate treatments for TB patients are increasing, which encourages continued transmission of TB. Therefore, the rapid detection of TB and drug resistance both optimizes treatment and improves outcomes and is also critical for reducing overall morbidity and mortality.
Traditionally, Mycobacterium tuberculosis (MTB) has been detected by Acid Fast Bacilli (AFB) smear and the gold standard microbial culture and identification [4,5]. Conventional drug susceptibility testing (DST) requires 3-8 weeks before the results are available, while rapid culture still takes an average of 7-9 days for smear-positive samples. Accordingly, there is current concern about the new molecular techniques that can detect the organism and detect also the antibiotic resistance either directly using respiratory samples or mycobacterial culture. These techniques based on detection of genetic mutations associated with drug resistance.
Although the molecular mechanisms of isoniazid resistance are not fully understood, numerous studies have linked them to distinct mutations in various genetic loci of the M. tuberculosis genome. Three loci most commonly affected are the katG gene, encoding catalase peroxidase, which transforms isoniazid to its pharmacologically active form, the inhA gene (with the mabAinhA promoter region), coding for enoyl-acyl carrier protein (ACP) reductase, a mycolic acid biosynthetic pathway enzyme [6-9], and the ahpC locus, including the structural gene of alkyl hydroperoxide reductase (ahpC), involved in the cellular response to oxidative stress, and the upstream regulatory region (oxyRahpC) [10-12]. Other genes that have been shown to be associated with isoniazid resistance include kasA, coding for b-ketoacyl ACP synthase, another enzyme involved in mycolic acid biosynthesis [13-15], ndh, coding for NADH dehydrogenase [16-18], and, more recently, nat and mshA, coding for arylamine N-acetyltransferase (NAT) [19], which, through acetylation of isoniazid, renders the drug therapeutically inactive, and the glycosyltransferase involved in the biosynthesis of mycothiol [19,20].
On the other hand, the cellular target of rifampicin is the betasubunit of bacterial DNA-dependent RNA polymerase, which is encoded by the rpoB gene. Point mutations in rpoB can render the organism resistant to RIF due to decreased binding affinity and can result in high-level resistance [21,22]. By targeting mutations in the 81-bp “core region” of the rpoB gene, more than 95% of all RMP resistant strains can be detected [23,24].
WHO-endorsed Genotype MTBDRplus assay has been validated and it is currently in use in several countries, providing appropriate results for drug resistance detection [25,26]. In the current study, the authors used Genotype MTBDRplus assay and specific gene sequencing to detect mutations associated with resistance to rifampicin and isoniazid in clinical isolates of M. tuberculosis.
Materials and Methods
Samples and Bacterial Isolates
During the year 2015, non-duplicated respiratory specimens were collected from 155 patients with chest symptoms, the samples collected from patients in TB screening canters in Cairo, Alexandria, Fayoum, Dakahlia and Jiza governorates in Egypt. All specimens were finally manipulated in Abassia Hospital-Cairo.
All specimens were routinely examined by Acid Fast Stain (AFB) staining followed by digestion using sputasol-sodium hydroxide [27]. Three drops of the decontaminated sediment were inoculated onto the LJ medium for each specimen. Newly inoculated Lowenstein–Jensen (LJ) media were kept in a launch position overnight. Subsequently, they were kept in the standby rack for 1-2 days to observe any gross contamination. All the cultures were examined for visible evidence of growth at daily intervals during the first week of incubation and were examined for visible evidence of growth at weekly intervals during the next 7-week incubation period. Cultures were regarded as negative only if there was no growth after 8 weeks of incubation.
Biochemical Reaction
The niacin and nitrate reduction tests were used for the primary identification of M. tuberculosis according to Kumar [28].
Antibiotic susceptibility and determination of minimum inhibitory concentration (MIC)
Conventional susceptibility testing (DST) was performed for Rifampicin (RIF) and Isoniazid (INH). It was performed using the proportion method on LJ Medium. MICs were defined as the lowest drug concentration after two-fold serially diluted concentration of the drugs that inhibits growth of more than 99.0% of a bacterial proportion of the tested M. tuberculosis within 14 to 21 days of incubation at 37°C.
For MIC detection, the following concentrations of anti-TB drugs were used in this study; 10, 20, 40, 125, 250, 500, 1000 and 2000 μg/ml for RIF; 0.02, 0.125, 0.250, 2.0 μg/ml for isoniazid INH. A strain was considered as MDR-TB if the cut off value more than 0.250 μg/ml (for isoniazid) & 40 μg/ml (for rifampicin).
Genotype MTBDRplus assay for drug-resistant Mycobacterium tuberculosis
Genotype MTBDRplus assay is a molecular genetic assay was used to detect resistant M. tuberculosis isolates to RIF and INH. The assay is based on the DNA-STRIP technology. The procedure includes a multiplex PCR amplification with biotinylated primers, and a reverse hybridization. The assay can detect the absence and/or presence of wild type (WT) and/or mutant (MUT) DNA sequences within specific region of the three target genes: rpoB gene (coding for the β-subunit of the RNA polymeraze), for the identification of rifampicin resistance; katG gene (coding for the catalase peroxidase), for isoniazid resistance; and the promoter region of inhA gene (coding for the NADH enoylACP reductase), for isoniazid resistance. The procedure was performed according to the manufacturer’s instructions (Hain Life sciences, Nehren, Germany).
Gene Sequencing
Sample preparation: A loopful of bacterial growth was transferred to glass bead tubes containing 3 ml of 0.9% NaCl from 6 weeks culture of M. tuberculosis. Bacteria were suspended for 15 min on vortex. Suspension was adjusted according to Mac Farland 1 to have 3 x 108 mycobacteria per 1 ml. Suspension was measured at 625 nm in spectrophotometer and adjusted so as to have an absorbance of 0.177. One ml of this suspension was mixed with 2 ml of 0.9% NaCl then 108 mycobacterial cells /ml suspension was obtained. From this main suspension 108, 104 and 101 dilutions were prepared by serial dilution. The dilutions were kept at –20 °C until used.
DNA extraction
The bacterial suspension used for inoculation was pelleted by centrifugation at 16,000 × g. Supernatants were decanted, and pelleted cells were re-suspended in 0.3 ml Tris-EDTA (TE) buffer and transferred to sterile 2-ml screw-cap tubes containing ∼250 μl of 0.1-mm-diameter glass beads (Becton Dickinson). Bacteria were heat killed at 80°C for 50 min and then frozen at −20°C; after thawing, the tubes were subjected to vortex mixing for 3 min and centrifuged for 5 min at 16,000 × g. The supernatant was transferred to a clean 2-ml tube for subsequent DNA purification using a DNA easy Blood and Tissue DNA extraction kit (Qiagen) as per the manufacturer's instruction.
PCR amplification and sequence analysis
For the detection of mutations associated with antibiotic resistance, PCR amplification was performed on randomly selected phenotypically resistant MTB and susceptible isolates. The primers used in the sequence analysis of rpoB, katG and mabinhA genes [29-31], are listed in Table 1.
| GenBank accession no | Product size | Primer sequence | Genes | Drugs |
|---|---|---|---|---|
| U12205 | 318 | 5'-CGATCACACCGCAGACGTTG-3' | rpoB | Rifampin |
| 5'-GGTACGGCGTTTCGATGAAC-3' | ||||
| X68081 | 232 | 5'-CATGAACGACGTCGAAACAG-3' | katG | Isoniazid |
| 5'-CGAGGAAACTGTTGTCCCAT-3' | ||||
| U66801.1 | 400 | 5'-ACATACCTGCTGCGCAAT-3' | mab-inhA | Isoniazid mab-inhA |
| 5'-TCACATTCGACGCCAAAC-3' |
Table 1: Primer sequences used for PCR amplification and sequence analysis.
A reaction mixture of 50 μl containing 5 μl of 10× PCR buffer, 1.5 mM MgCl2, 0.2 mM of deoxynucleotide triphosphate (dNTP), 0.2 μM of each primer, 2.5 U of Taq polymerase, and 5 μl of template DNA (10 ng) was prepared.
Amplification was performed on a thermal cycler (Bio-Rad, Hercules, CA, USA) with the following cycling program: initial denaturation at 95°C for 5 minutes followed by 30 cycles of denaturation at 95°C for 40 seconds, annealing at 64°C (rpoB, mab-inhA) at 55°C, (katG) for 1 minute, and extension at 72°C for 40 seconds, with a final extension at 72°C for 10 minutes. The PCR products were sent to on GATC Company, for purification and DNA sequencing (3730xl DNA Analyzers; Thermo Fisher Scientific).
Each sequence was analyzed with Bioedit and ClustalW software. Point mutations were identified by comparison with the sequence of the M. tuberculosis H37Rv reference strains in GenBank (NC 000962).
Statistical analysis
Data of this study were analyzed using statistical program for Social Science (SPSS) version 21.0. Quantitative data were expressed as frequency and percentage.
• The significance of Chi-square (X2) test was used in order to compare proportions between two qualitative parameters.
• The confidence interval was set to 95% and the accepted margin of error was set to 5%. The significance of Probability ( p-value) was evaluated as follow:
• P-value <0.05 was considered significant.
• P-value <0.001 was considered as highly significant.
• P-value >0.05 was considered insignificant.
Ethical Issues: The ethics committees in the participated hospitals had approved the study. All patients' information, data and individual test results were kept confidential.
Results
Patients and Samples
Among the 155 studied patients, 129 (83.2%) were males and remaining 26 (16.8%) were females. Most of the participants (87.7%) were more 31 years old (Table 2).
| Categories | Number of participants (N) | Frequency (%) |
|---|---|---|
| Sex | ||
| Male | 129 | 83.2 |
| Female | 26 | 16.8 |
| Age (years) | ||
| 0-15 | 6 | 3.9 |
| 16-30 | 13 | 8.4 |
| 31-45 | 48 | 31 |
| 46-60 | 54 | 34.8 |
| >60 | 34 | 21.9 |
Table 2: Characteristics of patients.
Acid Fast Bacilli (AFB) Microscopy
Microscopic examination revealed that 102 (65.8%) of 155 participants were positive for AFB. Based on the number of bacilli load positive specimens were categorized into five groups. Among the positive cases, 10 (9.8%) had 1-9 AFB/100 fields, 14 (13.7 %) were 1+, 32 (31.4 %) were 2+, 26 (25.5 %) were 3+ and 20 (19.6 %) were scanty positive (Table 3).
| AFB Microscopy | Number of cases (%) |
|---|---|
| AFB status (n=155) | |
| Negative | 53 (34.2%) |
| Positive | 102 (65.8%) |
| AFB grading (n=102) | |
| 1-9 AFB | 10 (9.8%) |
| 1+ | 14 (13.7) %) |
| 2+ | 32 (31.4) %) |
| 3+ | 26 (25.5) %) |
| Scanty | 20 (19.6) %) |
Table 3: Detection and grading of AFB in ZN stained sputum smears.
Mycobacterial Culture
Among 155 samples, 117 (75.5%) samples were found to be positive for culture, 35 (22.6%) samples were negative and 3 (1.9%) showed contamination on L-J.
Drug Susceptibility Testing (DST) and MIC Determination
After biochemical identification, drug susceptibility was performed using Lowenstein–Jensen medium-based proportion method for 106 identified M. tuberculosis complex isolates.
Minimum inhibitory concentration (MIC) was determined for 24 selected isolates of M. tuberculosis. Among these tested isolates 12, 5, 1 and 6 isolates were MDR, isoniazid resistant, rifampicin resistant and sensitive to both (isoniazid and rifampicin) respectively Tables 4-7.
| Rifampicin | Total | |||
|---|---|---|---|---|
| Sensitive (%) | Resistant (%) | |||
| Isoniazid | Sensitive (%) | 6 | 1 | 7 |
| Resistant (%) | 5 | 12 | 17 | |
| Total | 11 | 13 | 24 | |
Table 4: Drug susceptibility patterns among selected M. tuberculosis complex isolates.
| Variable | Rifampicin | P | Isoniazid | P | ||
|---|---|---|---|---|---|---|
| Sensitive | Resistant | Sensitive | Resistant | |||
| (N=11), n (%) | (N=13), n (%) | (N=7), n (%) | (N=17), n (%) | |||
| Patient age (years) | ||||||
| Dec-20 | 3 (27.3) | 4 (30.8) | >0.05 | 2 (28.6) | 5 (29.4) | >0.05 |
| >20-40 | 5 (45.5) | 4 (30.8) | 3 (42.9) | 6 (35.3) | ||
| >40-60 | 3 (27.3) | 5 (38.5) | 2 (28.6) | 6 (35.3) | ||
| Gender | ||||||
| Male | 9 (81.8) | 9 (69.2) | >0.05 | 6 (85.7) | 12 (70.6) | >0.05 |
| Female | 2 (18.2) | 4 (30.8) | 1 (14.3) | 5 (29.4) | ||
| Source Governorate | ||||||
| Alexandria | 3 (27.3) | 4 (30.8) | >0.05 | 2 (28.6) | 5 (29.4) | >0.05 |
| Cairo | 6 (54.5) | 5 (38.5) | 5 (71.4) | 6 (35.3) | ||
| Port Saied | 1 (9.1) | 1 (7.7) | 0 (0) | 2 (11.8) | ||
| Dakahlya | 1 (9.1) | 2 (15.4) | 0 (0) | 3 (17.6) | ||
| Giza | 0 (0) | 1 (7.7) | 0 (0) | 1 (5.9) | ||
| Treatment status | ||||||
| Relapsed | 1 (9.1) | 8 (61.5) | <0.001 | 1 (14.3) | 8 (47.1) | <0.05 |
| New case | 10 (90.9) | 0 (0) | 6 (85.7) | 4 (23.5) | ||
| Failed Treatment | 0 (0) | 5 (38.5) | 0 (0) | 5 (29.4) | ||
| HIV infection | ||||||
| Positive | 0 (0) | 3 (23.1) | >0.05 | 0 (0) | 3 (17.6) | >0.05 |
| Negative | 11 (100) | 10 (76.9) | 7 (100) | 14 (82.4) | ||
Note: N: Total number of sensitive or resistant isolates, n: number of sub-variable categories, p >0.05: not significant difference, p<0.05: significant difference, p <0.001: highly significant difference.
Table 5: Assessment of risk factors associated with drug resistance.
| Gene | Band | Gene Region of Mutation | MDR(n all = 12) n (%) | INH Mono resistant (n all=5) n (%) | RIF Mono resistant (n all=1) n (%) | INH/RIF Sensitive (n all =6) n (%) |
|---|---|---|---|---|---|---|
| katG | WT1 | 315 | 0 (0) | 1 (20) | 1(100) | 6 (100) |
| MUT1 | S315T1 | 12 (100) | 4 (80) | 0 (0) | 0 (0) | |
| MUT2 | S315T2 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| inhA | WT1 | 0.9375 | 11 (91.7) | 3 (60) | 1 (100) | 6 (100) |
| WT2 | -8 | 12 (100) | 5 (100) | 1 (100) | 6 (100) | |
| MUT1 | C15T | 1 (8.3) | 2 (40) | 0 (0) | 0 (0) | |
| MUT2 | A16G | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| MUT3A | T8C | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| MUT3B | T8A | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| rpoB | WT1 | 506-509 | 12 (100) | 5 (100) | 1 (100) | 6 (100) |
| WT2 | 510-513 | 12 (100) | 5 (100) | 1 (100) | 6 (100) | |
| WT3 | 513-517 | 12 (100) | 5 (100) | 1 (100) | 6 (100) | |
| WT4 | 516-519 | 12 (100) | 5 (100) | 1 (100) | 6 (100) | |
| WT5 | 518-522 | 12 (100) | 5 (100) | 1 (100) | 6 (100) | |
| WT6 | 521-525 | 12 (100) | 5 (100) | 1 (100) | 6 (100) | |
| WT7 | 526-529 | 12 (100) | 5 (100) | 1 (100) | 6 (100) | |
| WT8 | 530-533 | 0 (0) | 5 (100) | 1 (100) | 6 (100) | |
| MUT1 | D516V | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| MUT2A | H526Y | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| MUT2B | H526D | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| MUT3 | S531L | 8 (66.7) | 0 (0) | 0 (0) | 0 (0) |
Note: INH = Isoniazid; MDR = Multidrug resistant; RIF = Rifampicin
Table 6: Pattern of gene mutations in resistant M. tuberculosis complex isolates using the Genotype MTBDRplus assay.
| Drug/Isolate | MIC (µg/ml) (Interpretation) | Resistance Pattern | MTBDRplus mutation Pattern* | |
|---|---|---|---|---|
| INH | RIF | |||
| 66 | 2 (R) | >2000 (R) | MDR | ΔkatGWT1, katGMUT1, ΔrpoBWT8, rpoBMUT3 |
| 67 | >1.0 (R) | 250 (R) | MDR | ΔkatGWT1, katGMUT1, ΔrpoBWT8, rpoBMUT3 |
| 70 | >1.0 (R) | 250 (R) | MDR | ΔkatGWT1, katGMUT1, ΔrpoBWT8 |
| 72 | 2 (R) | 250 (R) | MDR | ΔkatGWT1, katGMUT1, ΔrpoBWT8, rpoBMUT3 |
| 75 | 2 (R) | 250 (R) | MDR | ΔkatGWT1, katGMUT1, ΔrpoBWT8, rpoBMUT3 |
| 79 | >1.0 (R) | 125 (R) | MDR | ΔkatGWT1, katGMUT1, ΔinhAWT1, inhAMUT1, ΔrpoBWT8 |
| 80 | >1.0 (R) | 125 (R) | MDR | ΔkatGWT1, katGMUT1, ΔrpoBWT8, rpoBMUT3 |
| 96 | >1.0 (R) | 250 (R) | MDR | ΔkatGWT1, katGMUT1, ΔrpoBWT8 |
| 626 | 0.250 (R) | 10 (S) | INHR | ΔkatGWT1, katGMUT1 |
| 669 | 0.125 (S) | 125 (R) | RIFR | WT |
| 701 | 2 (R) | 250 (R) | MDR | ΔkatGWT1, katGMUT1, ΔrpoBWT8, rpoBMUT3 |
| 740 | 0.250 (R) | 20 (S) | INHR | ΔkatGWT1, katGMUT1 |
| 742 | 0.125 (S) | 10 (S) | INHS/RIFS | WT |
| 747 | 2 (R) | 500 (R) | MDR | ΔkatGWT1, katGMUT1, ΔrpoBWT8, rpoBMUT3 |
| 769 | 2 (R) | 60 (R) | MDR | ΔkatGWT1, katGMUT1, ΔrpoBWT8 |
| 771 | 0.020 (S) | 30 (S) | INHS/RIFS | WT |
| 772 | 0.125 (S) | 20 (S) | INHS/RIFS | WT |
| 899 | 0.250 (R) | 20 (S) | INHR | ΔinhAWT1, inhAMUT1 |
| 943 | 0.125 (S) | 20 (S) | INHS/RIFS | WT |
| 944 | 0.125 (S) | 20 (S) | INHS/RIFS | WT |
| 945 | 0.020 (S) | 20 (S) | INHS/RIFS | WT |
| 946 | 0.250 (R) | 20 (S) | INHR | ΔkatGWT1, katGMUT1, ΔinhAWT1, inhAMUT1 |
| 948 | >1.0 (R) | >2000 (R) | MDR | ΔkatGWT1, katGMUT1, ΔrpoBWT8, rpoBMUT3 |
| 979 | 2 (R) | 30 (S) | INHR | ΔkatGWT1, katGMUT1 |
S: sensitive, R: resistant, MDR: multidrug resistant (resistant to both rifampicin and isoniazid), INH: isoniazid, RIF: rifampicin. Cut off concentrations are 0.250 µg/ml (for isoniazid) & 40 µg/ml (for rifampicin).
Δ: Missed band
Table 7: Assessment of risk factors associated with drug resistance.
Resistance Risk Factors
The patients’ age was categorized into 3 groups: 12-20, >20-40 and >40-60 years old. These groups had no significant impact on resistance to rifampicin and isoniazid (P value >0.05).
Similarity, patients’ gender, source governorate and patients’ infection with HIV had also no significant impact on resistance to rifampicin and isoniazid (P value >0.05).
On the other hand, treatment statuses of the patients were divided into relapsed, new cases and patients who had failed in treatment. Regarding rifampicin resistance, 8 patients among 13 patients (61.5%) infected with rifampicin resistant M. tuberculosis complex isolates were relapsed in their treatment, compared to 0 (0%) and 5 (38.5%) for new cases and failed treated patients respectively. While, 11 rifampicin sensitive isolates were isolated from 1 (9.1%) relapsed, 10 (90.9%) new cases and 0 (0%) patients failed in treatment (P <0.001).
Regarding isoniazid resistance, 8 patients among 17 patients (47.1%) infected with isoniazid resistant M. tuberculosis complex isolates were relapsed in their treatment, compared to 4 (23.5%) and 5 (29.4%) for new cases and failed treated patients respectively. While, 7 isoniazid sensitive isolates were isolated from 1 (14.3%) relapsed, 6 (85.7%) new cases and 0 (0%) patients failed in treatment (P <0.001) (Table 5).
Mutation Patterns Associated with RIF and INH Resistance using GenoType MTBDRplus Assay
The genotypic profile of resistance to RIF and NIH was examined by the Genotype MTBDRplus assay. In the 12 MDR isolates (based on MIC), the band of katG WT1 was missed accompanied by detection of the corresponding mutation band katG MUT2. Additionally, one isolate (8.35%) out of these 12 isolates missed also inhA WT2 band associated with appearance of inhA MUT1. Meanwhile, all these 12 isolates missed rpoB WT8 band and was associated in 8 (66.7%) of them by appearance of the mutation rpoB MUT3 band. The remaining four (33.3%) isolates (missed the rpoB WT8 band) did not show any corresponding mutations and can be considered as unknown (or non- inclusive).
For 5 isoniazid mono resistant isolates, 4 (80%) of them showed disappearance of the wild type band katG WT1 which was associated by the appearance of the corresponding mutation katG MUT1 band. Additional one of the mentioned 4 isolates and the fifth isolate (2/5, 40%) missed the wild type inhA WT1 while the mutation band “inhA MUT1” was observed. So, GenoType MTBDRplus assay could detect isoniazid resistance in 100% of these 5 isolates too. None of these 5 isolates showed any mutation regarding rpoB gene (no false positivity).
For one rifampicin mono resistant isolate, there were no detected mutations. The wild type bands for isoniazid resistance, namely katG WT1, inhA WT1 and inhA WT2 were preserved without appearance of any corresponding mutation bands.
None of the 6 INHS/RIFS isolates showed any mutation regarding katG, inhA or rpoB genes and accordingly no false positivity to antibiotic resistance was observed.
On the other hand the identification of 24/24 (100%) of M. tuberculosis complex isolates that were tested by GenoType MTBDRplus assay were molecularly confirmed based on the M. tuberculosis complex band included in the this assay.
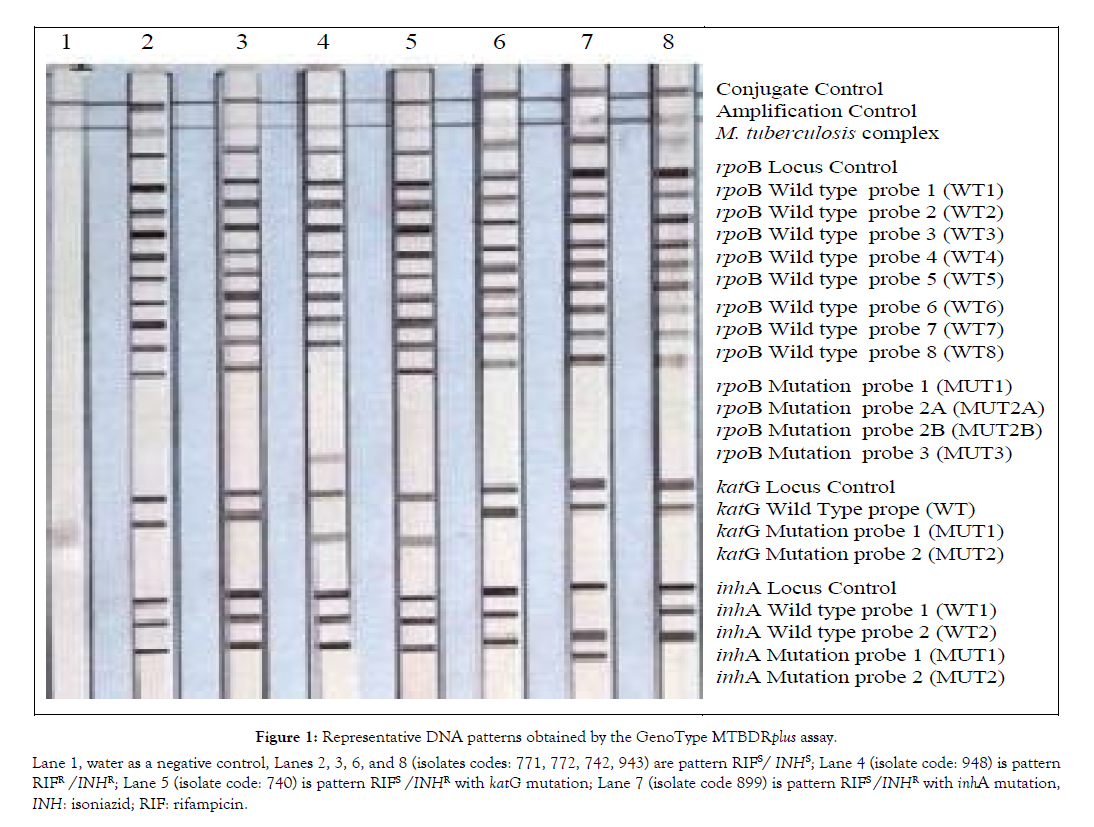
Table 7, also shows that all MDR had katG mutation associated with MIC for isoniazid ranged from Ë? 1.0 μg/ml to ≥ 2.0 μg/ml (as 2.0 μg/ml was the largest tested MIC). Among 5 strains of INHR, one strain had isoniazid MIC ≥ 2.0 μg/ml associated with katG mutation, four strains had isoniazid MIC 0.250 μg/ml associated with katG mutation (in two strains), katG, inhA mutations (1 strain) and only inhA mutation (1 strain). While 8 MDR strains had specific mutation in rpoB gene had MIC for rifampicin ranged from 125 μg/ml to ≥ 2000 μg/ml and 4 MDR strains had unknown mutation in rpoB gene had MIC for rifampicin ranged from 60 μg/ml to 250 μg/ml (Tables 6-8 and Figure 1).
Figure 1: Lane 1, water as a negative control, Lanes 2, 3, 6, and 8 (isolates codes: 771, 772, 742, 943) are pattern RIFS/ INHS; Lane 4 (isolate code: 948) is pattern RIFR /INHR; Lane 5 (isolate code: 740) is pattern RIFS /INHR with katG mutation; Lane 7 (isolate code 899) is pattern RIFS/INHR with inhA mutation, INH: isoniazid; RIF: rifampicin.
| RIF mutations | Frequency | INH mutations | Frequency | ||
|---|---|---|---|---|---|
| rpoB | MUT3 | 8 | katG | MUT1 | 14 |
| inhA | MUT1 | 1 | |||
| UK (WT8 missed) | 4 | katG +inhA | katG MUT1+inhA MUT1 | 2 | |
| Total RIF resistant | 12 | Total INH resistance | 17 | ||
| (MDR+mono RIF) | (MDR+mono INH) | ||||
Note: INH: isoniazid; RIF: rifampicin; UK: unknown mutation characterized by no hybridization to one or more wild-type probes nor to any of mutation probes; WT: wild type
Table 8: Frequency of gene mutations in resistant Mycobacterium tuberculosis complex isolates using the GenoType MTBDRplus assay.
Sequencing of drug resistance genes
Gene sequencing of eight isolates with discordant DST results showed that, it could detect 100% of resistance among the tested isolates. Mutations were detected in codon 315 of katG, position –15 (C-T) of the inhA promoter, Codon 531 of rpoB gene (Table 9).
| Target resistance gene for sequencing | Phenotypic resistance pattern | Number of tested isolates | Detected mutation by MTBDRplus | Detected mutations by PCR based-gene Sequencing |
|---|---|---|---|---|
| katG | MDR | 2 | Missed katG WT1 & | codon 315 of katG |
| katG MUT1 | ||||
| katG | mono INHR | 2 | Missed katG WT1 & | codon 315 of katG |
| katG MUT1 | ||||
| inhA | mono INHR | 1 | Missed inhA WT1 & inhA MUT1 | position –15 (C-T) of the inhA promoter |
| rpoB | MDR | 2 | Missed rpoB WT8 & rpoB MUT3 | Codon 531 of rpoB |
| rpoB | mono RIFR | 1 | No specific mutation | Codon 531 of rpoB |
Note: MDR: multidrug resistant, INHR: isoniazid resistant, RIFR: rifampicin resistant
Table 9: Frequency of gene mutations in selected isolates of resistant Mycobacterium tuberculosis complex using the gene sequencing.
Discussion
The study showed that most of rifampicin resistant cases (61.5%) were from patients relapsed in treatment and nothing from new cases, but 90.9% of rifampicin sensitive isolates were from new cases. Similarly, 47.1% of isoniazid resistant isolates were from patients relapsed in treatment while 85.7% of isoniazid sensitive isolates were from new patients. This figure shows the association between drug resistance and relapse in treatment. Relapsed patient is defined as TB patient who declared cured or completed treatment, but reported back to health service and is now found to be sputum smear positive [29,30]. But which is the result of the other, the treatment relapse or drug resistance. Jamieson (2014) [31] stated that relapse risk factors included initial drug resistance and drug irregularity. It may be logic that hetero-resistance path of the bacteria may lead to selection of resistant strains after treatment coarse and reappearance (relapse treatment) of infection by these resistant strains. The current study findings is also in agreement with a previous WHO report in specific countries that detected MDR-TB in 28% and 62% of new patients and previously treated patients respectively (WHO, 2010) [32]. As an example from Cameron, Jiang et al., (2012) reported that MDR-TB strains were 24.9% and 8.9% among previously treated and new patients respectively. Global Tuberculosis Report (2019) [1,33], mentioned that Globally, 3.5% (2018 report) then 3.4% (2019 Report) of new TB cases and 18% of previously treated cases had MDR/RR-TB (Multi-drug resistant/Rifampicin resistant TB).
On the other hand, patient age, gender, geographic location and even HIV status (a well-known risk factor for TB infection) were not considered as significant risk factors for drug resistance in this study. However, a review of possible TB-drug resistance risk factors was performed by Kelley (1997) [34] who mentioned that these risk factors include diabetes mellitus, error in drug regimen, dosing interval and duration, failure to identify pre-existing resistance, variations in bioavailability of anti-TB drugs and non-adherence to prescribed treatment. However, changes in the genomic content are a major underlying event which is associated with the emergence of resistant variants. The Male: Female ratio of the TB cases incident for all ages ranged from 1.1 in the WHO Eastern Mediterranean Region to 2.1 in the Western Pacific region [1] which has no impact on drug resistance variation between males and females in the current study. The current results are also in agreement with Lempens et al., [35] who revealed that gender is not an associated risk factor for MDR-TB while previous TB treatment is a risk factor but in contrast to the current study, they found that younger age group (less than 25 years old) was also a risk factor, which may be affected by secondary risk factors like social behaviors, drug abuse or other factors related to young age that are different from one community to another.
Many studies reviewed the co-existence of HIV and TB infections, the susceptibility of HIV/AIDS patients to TB infections or low CD4 cells count as a risk factor for TB infection [1,36-38]. But regarding TB drug resistance, the current study agreed with Perizzolo et al., [39], that assessed the impact of HIV/TB co-infection on drug resistance emergence and reported that HIV/TB co-infection did not significantly affect the mutation rate or emergence of resistance in M. tuberculosis within patients. On the other hand Singh et al., stated that coinfection of HIV and M/XDR-TB is responsible for all forms of M/XDR-TB epidemics or outbreaks [40].
In response to the emergence of MDR M. tuberculosis, improvement of new molecular techniques is always needed; these techniques can screen a wide range of drug resistance related genetic markers. These techniques will also help in rapid and accurate diagnosis, reporting and treatment of TB patients and especially patients suffering from MDR-TB. The Global Tuberculosis Report (2019) [1] stated that there are significant gaps between the estimated number of new cases and the number actually reported due to underreporting of cases, and underdiagnosed cases. It mentioned an example of Indonesia, in 2017, in a national study found that about 20% of new cases were not diagnosed; of the approximately 80% of new cases that were detected, 41% were not reported. Global Tuberculosis Report (2019) [1] reported some progress in testing, detection and treatment of MDR/RR-TB between 2017 and 2018. Globally in 2018, 51% of people with bacteriologically confirmed TB were tested for rifampicin resistance, up from 41% in 2017.
Some techniques that have been evaluated by different authors include Genotype MTBDR against Genotype MTBDRplus, multiplex ligaton-dependent probe amplification (MLPA) and/or Genotype® MTBDRplus, and many others [39-43]. The current study revealed again the validity of Genotype MTBDRplus assay as rapid detector of MDR M. tuberculosis isolates which improves care of the patients and treatment outcome. WHO generally encourages the rapid molecular techniques and according to its review in August 2019, many new techniques are under development including Gendrive MTB/RIF ID, Epistem, UK & Xpert XDR-TB cartridge, Cepheid, USA & TruArray MDR-TB, Akkoni, USA & INFINITIMTB Assay, AutoGenomics, USA & FluoroType XDR-TB assay, Hain Lifescience, Germany & MeltPro TB assay, Zeesan Biotech, China and QuantuMDx, POC, UK [1,2].
All isoniazid resistant strains were detected by GenoType plus assay and it could clearly detect rifampicin resistance among 66.7% of MDR isolates that showed mutation band rpoB MUT3 while 33.3% of them did not show any mutation bands (although the wild rpoB WT8 was missed) and can be considered as unknown (or non-inclusive). One mono-resistant rifampicin isolate did not show rifampicin mutation band by GenoType plus assay, but it showed mutation in Codon 531 of rpoB as detected by sequence analysis which is unexpected result. Sequencing is considered as a reference method as it used as a tool to test the clinical validity of the new molecular assays, in addition to reference standards of culture, phenotypic DST and Xpert® MTB/ RIF [1,17]. Accordingly, rifampicin resistance in this strain could be associated with mutation in Codon 531 of rpoB (based on molecular sequencing) and in this case Genotype MTBDR plus assay could not detect the associated mutation as MUT3 band was absent and WT8 band was preserved. If the results of Genotype MTBDRplus assay and sequencing are combined, this strain could have hetero-resistance pattern, which might be due to selection of cells with random mutations during inadequate treatment or mixed infections with sensitive and resistant cells [44,45]. The hetroresistance phenomenon may affect the effective treatment of patients and lead to the development of drug resistance. But the current result may reflect also low sensitivity or technical error of the MTBDRplus assay during testing of this strain. Some other authors reported also similar observations like the detection of codon 531 in MDR-TB strains by sequencing with no corresponding mutation detected by Genotype MTBDRplus assay [46,47] or with only disappearance of the wild type band, without specific mutation band by Genotype MTBDRplus assay [48,49]. The last case is also the same observed in 4 strains of MDR-TB in the current study.
The non-detected mutations by MTDRplus technique in some resistant isolates is explained as the known molecular determinants cannot detect 100% of the drug resistant isolates.
Haile et al., 2020 reported that Genotype MTBDRplus assay demonstrated a mutation of only 5.4% and 1.1% for isoniazid and rifampicin respectively [50], while Dean et al., (2020) found high confidence resistance conferring mutations were identified in 78.6% of isolniazid resistant isolates [51]. This also may be due to the presence of some strains exhibited heteroresistance as described by other researchers on rpoB, D516V/S531L [52,53]. Gene sequencing of eight selected isolates, with determined mutations or missed wild type band by MTDRplus technique, could detect resistance mutations mainly in codon 315 (katG gene), position -15 in inhA promoter gene) associated with isoniazid resistance and codon 531 (rpoB gene) associated with rifampicin resistance. In previous studies also the authors have identified highly variable frequencies of these mutations; with katG315 mutations accounting for 42 to 95% and inhA-15 mutations accounting for 6 to 43% of phenotypic INH resistance [53,54]. A more recent study in Kyrgyz Republic, revealed that the most prevalent mutation in RIF-resistant isolates was found in code 531 of rpoB gene and code 315 in katG gene of INH-resistant M. tuberculosis [34,46]. Actually, they detected 13 mutations in 7 codons of rpoB gene in RIF-resistant samples (n=185), where the most prevalent locations were codons 531 (64.8%), 526 (17.3%), 516 (8.1%), and 511 (5.4%), whilst 513, 533 and 522 codons mutations were quite rare, altogether explaining only 4.4% of all rpoB gene mutations. While, mutation in codon 315 prevailed in katG gene with three mutation variants. Similarly, for isoniazid, Ullah et al., [51], reported that katG and inhA promoter mutations can be considered as predictive markers of isoniazid resistance. Testing only for katG 315 and inhA –15 mutations detected isoniazid resistance in 84% of multi-drug resistant M. tuberculosis isolates.
Generally genotyping techniques allows distinguishing between recurrent cases of reinfection or reactivation [54,55], it supports epidemiological studies to monitor genotypic diversity of M. tuberculosis strains, giving that particular genotypes are correlated with bacterial virulence, disease dynamics and outbreaks. Although the efficiency of MTBDRplus assay for early detection of M. tuberculosis with rifampicin and/or isoniazid resistance. There are some challenges such as presence of the wild type cannot exclude the resistance to other antibiotics and should even be confirmed for susceptibility to rifampicin and/or isoniazid [55,56]. In the current study all tested MDR strains have katG gene mutation which according to many authors such as Vilche et al., [57], likely to be associated with high resistance level to isoniazid. Zhang et al., [58], also used GenoType MTBDRplus assay for identification of the katG as predictor for high level INH resistance and inhA for low level INH resistance. On the other hand, Lempens et al., [35] reported that isoniazid high level of resistance was associated with combined mutations in katG and inhA genes but isolates harbored mutations in inhA gene alone expressed moderate level of resistance and they concluded that line probe assay is not sufficiently accurate for prediction of such resistance level. Machado et al. [36] also reported that strain specific factors may be involved as strains from Lisboa family was associated with high level of isoniazid resistance in presence of mutation in the inhA regulatory region together with mutation in inhA coding region in contrast to the documented association between inhA mutation and low level isoniazid resistance [59]. MIC for isoniazid in the current study ranged from ˃ 1.0 μg/ ml to 2.0 μg/ml or more among MDR isolates and one strain of INHR associated with katG gene mutation while in other INHR strains, MIC was 0.250 μg/ml associated with katG mutation, katG, inhA mutations and only inhA mutation. This may conclude that katG mutation predicts isoniazid MIC of more 1.0 μg/ml among MDR strains in the tested strains. Huyen et al., [26] concluded that katG315 but not inhA mutation associated with unfavorable treatment outcome including patient relapse and death which agrees with the current study as among 17 isoniazid resistant isolates (16 isolates of them harbored katG mutation), 76.5% were either relapse or failed in treatment.
It has been reported that mutations in the codons 516, 526 and 531 in rpoB are associated with high level (70% to 95%) of rifampicin resistance [32]. Jamieson et al., [31] reported some association between rpoB and high level rifampicin resistance but recommended the combination between phenotypic and molecular methods for testing rifampicin susceptibility and the current study agrees with this conclusion, as specific mutation in rpoB gene was associated with higher levels of rifampicin resistance than that associated with unknown mutations by MTBDRplus assay among MDR isolates which may give some hint about resistance level.
Conclusion and Recommendation
As a conclusion, Moringa oleifera contains polyphenol compounds in all extract (aqueous, ethanol, ethyl acetate and chloroform) have antioxidant, anticancer and anti-inflammatory activity, also all extracts of moringa have antibacterial activities.
Acknowledgement
The molecular techniques are able initially to predict rifampicin and isoniazid drug resistance and also level of resistance, but combination between molecular and phenotypic methods still recommended.
Heteroresistance pattern and mixed infection with additional non-tuberculous mycobacteria are still considerable challenges. Effort should continue to explore more sensitive, rapid and specific molecular techniques for detection of resistance in M. tuberculosis, which will also improve the treatment opportunities. This effort is already encouraged, supported, evaluated and planned by WHO. In 2020, the final draft of the global strategy on TB research and innovation was presented for consideration by the 145th session of the WHO Executive Board (in January) and will be presented to the 72nd World Health Assembly (in May).
REFERENCES
- https://www.who.int/tb/publications/global_report/en/
- Abanda NN, Djieugoué JY, Lim E, Pefura-Yone EW, Mbacham WF, Vernet G, et al. Diagnostic accuracy and usefulness of the Genotype MTBDRplus assay in diagnosing multidrug-resistant tuberculosis in Cameroon? A cross-sectional study. BMC Infectious Diseases. 2017;17(1):379.
- Afanasev MV, Ikryannikova LN, Ilina EN, Sidorenko SV, Kuzmin AV, Larionova EE, et al. Molecular characteristics of rifampicin- and isoniazid-resistant Mycobacterium tuberculosis isolates from the Russian Federation. J Antimicrob Chemother. 2007;59(6):1057-1064.
- Ahmad S, Mokaddas E. Recent advances in the diagnosis and treatment of multidrug-resistant tuberculosis. Respir Med. 2009;103(12):1777-1790.
- Alelign A, Zewude A, Mohammed T, Tolosa S, Ameni G, Petros B. Molecular detection of Mycobacterium tuberculosis sensitivity to rifampicin and isoniazid in South Gondar Zone, northwest Ethiopia. BMC Infect Dis. 2019;19(1):343.
- Ando H, Kondo Y, Suetake T, Toyota E, Kato S, Mori T, et al. Identification of katG Mutations Associated with High-Level Isoniazid Resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2010;54(5):1793-1799.
- Bakonyte D, Baranauskaite A, Cicenaite J, Sosnovskaja A, Stakenas P. Molecular characterization of isoniazid-resistant Mycobacterium tuberculosis clinical isolates in Lithuania. Antimicrob Agents Chemother. 2003;47(6):2009-2011.
- Cardoso RF, Cardoso MA, Leite CQ, Sato DN, Mamizuka EM, Hirata RD, et al. Characterization of ndh gene of isoniazid resistant and susceptible Mycobacterium tuberculosis isolates from Brazil. Mem Inst Oswaldo Cruz. 2007;102:59-61.
- Cavusoglu C, Hilmioglu S, Guneri S, Bilgic A. Characterization of rpoB mutations in rifampin resistant clinical isolates of Mycobacterium tuberculosis from Turkey by DNA sequencing and line probe assay. J Clin Microbiol. 2002;40(12):4435-4438.
- Chen X, Kong F, Wang Q, Li C, Zhang J, Gilbert GL. Rapid detection of Isoniazid, Rifampin, and Ofloxacin Resistance in Mycobacterium tuberculosis clinical isolates using high-resolution melting analysis. J Clin Microbiol. 2011;49(10):3450-3457.
- Coelho MB, Costa ER, Vasconcellos SE, Linck N, Ramos RM, Amorim HL, et al. Sequence and structural characterization of tbnat gene in isoniazid-resistant Mycobacterium tuberculosis: Identification of new mutations. Mutat Res. 2011;712:33-39.
- Dalla Costa ER, Vasconcelos SEG, Esteves LS, Gomes HM, Gomes LL, Silva PA, et al. Multidrug-resistant Mycobacterium tuberculosis of the Latin American Mediterranean lineage, wrongly identified as Mycobacterium pinnipedii (spoligotype international type863 [SIT863]), causing active tuberculosis in South Brazil. J Clin Microbiol. 2015;53:3805-3811.
- Dheda K, Chang KC, Guglielmetti L, Furin J, Schaaf HS, Chesov D, et al. Clinical management of adults and children with MDR and XDR-TB. Clin Microbiol Infect. 2016;23:131-140.
- dos Santos PFG, Costa ER, Ramalho DM, Rossetti M, Barcellos R, Nunes L de S, et al. Detection of tuberculosis drug resistance: a comparison by Mycobacterium tuberculosis MLPA assay versus GenoType® MTBDRplus. Mem Inst Oswaldo Cruz, Rio de Janeiro. 2017;112(6):396-403.
- Eldholm V, Rieux A, Monteserin J, Lopez JM, Palmero D, Lopez B, et al. Impact of HIV co-infection on the evolution and transmission of multidrug-resistant tuberculosis. eLife. 2016;5:e16644.
- Forth Global Report 2008. World Health Organization and International Union against Tuberculosis and Lung Disease (WHO/IUATLD). Global Project on Anti-Tuberculosis Drug Resistance Surveillance 2002-2007: Anti-Tuberculosis Drug Resistance in the World: Geneva, Switzerland, WHO.
- Gandhi NR, Nunn P, Dheda K, Schaaf HS, Zignol M, van Soolingen D, et al. Multidrug-resistant and extensively drug-resistant tuberculosis: A threat to global control of tuberculosis. Lancet. 2010;375(9728):1830-1843.
- Global tuberculosis report 2018. Geneva: World Health Organization; License: CC BY-NC-SA 3.0 IGO
- Gomes HM, Elias AR, Oelemann MA, Pereira MA, Montes FF, Marsico AG, et al. Spoligotypes of Mycobacterium tuberculosis complex isolates from patients residents of 11 states of Brazil. Infect Genet Evol. 2012;12(4):649-656.
- Gulrez SA. DOTS for TB relapse in India: A systemic review. Lung India, 2012;29(2):147-153.
- Guo H, Seet Q, Denkin S, Parsons L, Zhang Y. Molecular characterization of isoniazid resistant clinical isolates of Mycobacterium tuberculosis from the USA. J Med Microbiol. 2006;55(11):1527-1531.
- Hazbon MH, Brimacombe M, Bobadilla del Valle M, Cavatore M, Guerrero MI, Varma-Basil M, et al. Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2006;50(8):2640-2649.
- Herrera L, Valverde A, Saiz P, Sáez-Nieto JA, Portero JL, Jiménez MS. Molecular characterization of isoniazid-resistant Mycobacterium tuberculosis clinical strains isolated in the Philippines. Int J Antimicrob Agents. 2004;23(6):572-576.
- Hillemann D, Rusch-Gerdes S, Richter E. Evaluation of the GenoType MTBDRplus Assay for Rifampin and Isoniazid Susceptibility Testing of Mycobacterium tuberculosis Strains and Clinical Specimens. J Clin Microbiol. 2007;45(8):2635-2640.
- Hoek KG, Van Rie A, van Helden PD, Warren RM, Victor TC. Detecting drug resistant tuberculosis: The importance of rapid testing. Mol Diagn Ther. 2011;15(4):189-194.
- Huyen MNT, Cobelens FGJ, Buu TN, Lan NTN, Dung NH, Kremer K, et al. Epidemiology of isoniazid resistance mutations and their effect on tuberculosis treatment outcomes. Antimicrob Agents Chemother. 2013;57(8):3620-3627.
- Yuen K-Y, Yam W-C, Wong L-P, Seto W-H. Comparison of Two Automated DNA Ampliï¬Âcation Systems with a Manual One-Tube Nested PCR Assay for Diagnosis of Pulmonary Tuberculosis. J Clin Microbiolol. 1997;35(6):1385-1389.
- Kumar S. Mycobacterium tuberculosis: in, Textbook of Microbiology, Jaypee Brothers Medical Publishers, India, 2012.
- Isakova J, Sovkhozova1 N, Vinnikov D, Goncharova Z, Talaibekova E, Aldasheva N, et al. Mutations of rpoB, katG, inhA and ahp genes in rifampicin and isoniazid-resistant Mycobacterium tuberculosis in Kyrgyz Republic. BMC Microbiology. 2018;18(1):22.
- Jagielski T, Bakuła Z, Roeske K, Kamin´ski M, Napio ´rkowska A, Augustynowicz-Kopec E, et al. Detection of mutations associated with isoniazid resistance in multidrug-resistant Mycobacterium tuberculosis clinical isolates. J Antimicrob Chemother. 2014;69(9):2369-2375.
- Jamieson FB, Guthrie JL, Neemuchwala A, Lastovetska O, Melano RG, Mehaffya C. Profiling of rpoB Mutations and MICs for Rifampin and Rifabutin in Mycobacterium tuberculosis. J Clin Microbiol. 2014;52(6):2157-2162.
- World Health Organization. Multidrug and Extensively Drug-Resistant TB. Global Report of Surveillance and Response. Geneva, Switzerland; WHO Press. 2010.
- Jiang LJ, Wu WJ, Wu H, Ryang SS, Zhou J, Wu W, et al. Rapid detection and monitoring therapeutic efficacy of Mycobacterium tuberculosis complex using a novel real-time assay. J Microbiol Biotechnol. 2012;22(9):1301-1306.
- Kelley CL, Rouse DA, Morris SL. Analysis of ahpC gene mutations in isoniazid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1997;41(9):2057-2068.
- Lempens P, Meehan CJ, Vandelannoote K, Fissette K, de Rijk P, Van Deun A, et al. Isoniazid resistance levels of Mycobacterium tuberculosis can largely be predicted by highconfidence resistance-conferring Mutations. Scientific Reports. 2018;8(1):3246.
- Machado D, Perdiga˜o J˜o, Ramos J, Couto I, Portugal I, Ritter C, et al. High-level resistance to isoniazid and ethionamide in multidrug-resistant Mycobacterium tuberculosis of the Lisboa family is associated with inhA double mutations. J Antimicrob Chemother. 2013;68(8):1728-1732.
- Narvskaya O, Otten T, Limeschenko E, Sapozhnikova N, Graschenkova O, Steklova L, et al. Nosocomial outbreak of multidrugresistant tuberculosis caused by a strain of Mycobacterium tuberculosis W-Beijing family in St. Petersburg, Russia. Eur J Clin Microbiol Infect Dis. 2002;21(8):596-602.
- Nikolayevsky V, Balabanova Y, Simak T, Malomanova N, Fedorin I, Drobniewski F. Performance of the Genotype® MTBDRplus assay in the diagnosis of tuberculosis and drug resistance in Samara, Russian Federation. BMC Clin Pathol. 2009;31:1381-1387.
- Perizzolo PF, Dalla Costa ER, Ribeiro AW, Spies FS, Ribeiro MO, Dias CF, et al. Characteristics of multidrug-resistant Mycobacterium tuberculosis in Southern Brazil. Tuberculosis (Edinb). 2012;92(1):56-59.
- Singh A, Prasad R, Balasubramanian V, Gupta N. Drug-Resistant Tuberculosis and HIV Infection: Current Perspectives. HIV AIDS (Auckl). 2020;12:9-31.
- Pozzi G, Meloni M, Iona E, Orru G, Thoresen OF, Ricci ML, et al. rpoB Mutations in Multidrug-Resistant Strains of Mycobacterium tuberculosis Isolated in Italy. J Clin Microbiol. 1999;37:1197-1199.
- Ramaswamy SV, Reich R, Dou SJ, Jasperse L, Pan X, Wanger A, et al. Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2003;47:1241-1250.
- Rattan A, Kalia A, Ahmad N. Multidrug resistant Mycobacterium tuberculosis: molecular perspectives. Emerg Infect Dis. 1998;4(2):195-209.
- Rossetti ML, Valim AR, Silva MS, Rodrigues VS. Resistant tuberculosis: a molecular review. Rev Saude Publica. 2002;36(4):525-532.
- Rumende CM. Risk factors for multidrug-resistant tuberculosis. Act Med Indones. 2018;50(1):1-2.
- Schön T, Miotto P, Ksoser CU, Viveiros M, Bottger E, Cambau E. Mycobacterium tuberculosis drug-resistance testing: Challenges, recent developments and perspectives. Clin Microbiol Infect. 2017;23(3):154-160.
- Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30(12):e57.
- Sethi S, Singh S, Dhatwalia SK, Yadav R, Mewara A, Singh M, et al. Evaluation of in-house loop-mediated isothermal amplification (LAMP) assay for rapid diagnosis of M. tuberculosis in pulmonary specimens. J Clin Lab Anal. 2013;27(4):272-276.
- Silva MSN, Senna SG, Ribeiro MO, Valim AR, Telles MA, Kritski A, et al. Mutations in katG, inhA, and ahpC Genes of Brazilian Isoniazid-Resistant Isolates of Mycobacterium tuberculosis. J Clin Microbiol. 2003;41(9):4471-4474.
- Haile B, Tafess K, Zewude A, Yenew B, Siu G, Ameni G. Spoligotyping and drug sensitivity of Mycobacterium tuberculosis isolated from pulmonary tuberculosis patients in the Arsi Zone of southeastern Ethiopia. New Microbe and New Infect. 2020;33:1-8.
- Dean AS, Zignol M, Cabibbe AM, Falzon D, Glaziou P, Cirillo DM, et al. Prevalence and genetic profiles of isoniazid resistance in tuberculosis patients: A multicountry analysis of cross-sectional data. PLoS Med 2020;17(1):e1003008.
- Simons SO, van der Laan T, de Zwaan R, Kamst M, van Ingen J, Dekhuijzen PN, et al. Molecular drug susceptibility testing in the Netherlands: Performance of the MTBDRplus and MTBDR assays. Int J Tuberc Lung Dis. 2015;19(7):828-833.
- Sirous M, Khosravi AD, Tabandeh MR, Salmanzadeh S, Ahmadkhosravi N, Amini S. Molecular Detection of Rifampin, Isoniazid, and Ofloxacin Resistance in Iranian Isolates of Mycobacterium tuberculosis by High-resolution Melting Analysis. Infect Drug Resist. 2018;11:1819-1829.
- Ullah I, Javaid A, Tahir Z, Ullah O, Shah AA, Hasan F, et al. Pattern of drug resistance and risk factors associated with development of drug resistant Mycobacterium tuberculosis in Pakistan. PLoS ONE 2016;11(1): e0147529.
- Unis G, Ribeiro AW, Esteves LS, Spies FS, Picon PD, Dalla Costa ER, et al. Tuberculosis recurrence in a high incidence setting for HIV and tuberculosis in Brazil. BMC Infect Dis. 2014;14:548.
- van Deun A, Barrera L, Bastian I, Fattorini L, Hoffmann H, Kam KM, et al. Mycobacterium tuberculosis strains with highly discordant rifampin susceptibility test results. J Clin Microbiol. 2009;47(11):3501-3506.
- Vilcheze C, Wang F, Arai M, Hazbón MH, Colangeli R, Kremer L, et al. Transfer of a point mutation in Mycobacterium tuberculosis inhA resolves the target of isoniazid. Nat Med. 2006;12:1027-1029.
- Zhang M, Yue J, Yang YP, Zhang HM, Lei JQ, Jin RL, et al. Detection of mutations associated with isoniazid resistance in Mycobacterium tuberculosis isolates from China. J Clin Microbiol. 2005;43:5477-5482.
- Ziemlé CM, Issiaka S, Olivier WCS, Daouda M, Donald EM, Yi-Ming AC. Risk factors of tuberculosis infection among HIV/AIDS patients in Bukina Faso. AIDS Res Hum Retroviruses. 2013;29(7):1045-1055.
Citation: Hosny AM, Shady HMA, Essawy AKE (2020) rpoB, katG and inhA Genes: The Mutations Associated with Resistance to Rifampicin and Isoniazid in Egyptian Mycobacterium tuberculosis Clinical Isolates. J Microb Biochem Technol.12:3. doi: 10.35248/1948-5948.20.12.428
Copyright: © 2020 Hosny AM et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Competing interests: The authors have declared that no competing interests exist.