Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- CiteFactor
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Review Article - (2021) Volume 12, Issue 6
Recent Advances in Potato Late Blight Disease Management Strategies
Yitagesu Tadesse*, Dereje Amare and Asela KeshoReceived: 25-May-2021 Published: 28-Jun-2021, DOI: 10.35248/2157-7471.21.12.559
Abstract
Potato is the world’s number one non-grain food commodity, with production reaching 325 million tons. Some inherent qualities give potato a competitive edge over the leading food crops. It is able to produce more protein and carbohydrates per unit area than cereals. Among all the crops grown worldwide, is known to suffer the greatest losses from disease attack. Among those diseases late blight and bacterial wilt are the most economical diseases which incurred 100% yield loss. The main Objective of this review was to understand very well the recent advances on potato late blight disease management up to gene for gene level and incorporate these recent molecular advances with the conventional one for mitigating the virulence spectrum of the pathogen. The population of P. infestans characterized using molecular markers has led to better understanding of pathogen at molecular level. Mitochondrial DNA haplotyping of P. infestans has revealed that mt Ia is displacing the other haplotypes globally at a faster rate.
Keywords
Recent advance; P. infestans; Epidemiology; Genetic variation; Restriction fragment length polymorphism; Interaction transcriptome; PCR-based methods
Introduction
Potato (Solanum tuberosum L.) is the fourth major crop of the world after rice, wheat and maize [1]. It is used as vegetable, stock feed and in industries for manufacturing starch, alcoholic beverages and other processed products. It provides essential body building substances such as proteins, vitamins, minerals (P, Ca, Mg, K, Fe, S, Cl) [2]. It is an important food source globally. It is cultivated worldwide in over one hundred countries throughout Africa, Asia, Australia, Europe, and North and South America [3]. Some inherent qualities give potato a competitive edge over the leading food crops. In fact, it is able to produce more protein and carbohydrates per unit area than cereals and some leguminous crops like soybeans [4]. It is the world’s number one non-grain food commodity, with production reaching 325 million tons in 2007 [1]. Its production in developed countries, especially in Europe and Commonwealth independent states, has been declining on average by one percent per annum over the past 20 years. However, the output in developing countries has been expanding at an average rate of 5% per year [1]. In Ethiopia, potato production is expanding steadily. The total area under potato crop in 2012 was estimated to be 54,007 hectares [5]. According to FAO [1] production has increased from 280,000 tons in 1993 to around 525,000 tons in 2007 and during 2007 production year the total area covered with potato was 73,095 hectares, while the yield was 7.2 tons/hectare. It has a promising prospect in improving the quality of the basic diet in both rural and urban areas of the country. The protein from potato is a good composition with regard to essential amino acids in human nutrition. Potato also has substantial amount of vitamins, minerals and trace elements. Such crop undoubtedly is very important for Ethiopia, where adequate protein and supplies of calories are the apparent nutritional problems [6].
Among all the crops grown worldwide, is known to suffer the greatest losses from disease attack. Its cultivation is constrained by several biotic factors such as Early blight, late blight, wart, black scurf, charcoal rot, powdery scab, Fusarium wilt, Verticillium wilt, Sclerotium wilt, common scab, Brown rot, soft rot, potato virus X, potato virus Y, crinkle mosaic, potato acuba mosaic virus and witch’s broom, of which late blight caused by Phytophthora infestans (Mont. de Bary) is considered the most important, highly devastating disease [7]. It is become endemic in potato growing areas, with an incidence ranging from 50-100 percent [8]. Late blight of potato, caused by P. infestans (Mont. De Bary), is among its most important diseases, being especially devastating in the major potato growing tropical highlands of Sub-Saharan African. In the mid-1840s, a devastating potato disease swept continental Europe, the British Isles and Ireland. It is estimated that Ireland, as a direct consequence of late blight, lost more than a quarter of its 8 million inhabitants to starvation and emigration, making this one of the most significant crop diseases in history. However, it was not until 1876 that a micro-organism named Phytophthora (meaning ‘plant destroyer’) infestans was conclusively demonstrated to be responsible for potato late blight [9]. In the mid 1800s, late blight caused widespread crop failures throughout Northern Europe including Ireland where it was responsible for the Irish famine [10]. Since then, it has spread far and wide and now occurs wherever potatoes are grown. Losses due to P. infestans have been estimated to € 12 billion per annum of which the losses in developing countries have been estimated around € 10 billion per annum [11].
P. infestans is heterothallic oomycete with two mating types, A1 and A2, which evolved in the Toluca valley, central Mexico. Until the 1980s the A2 mating type was confined to Mexico, previous spread of the pathogen being attributed to a single A1 isolate of the pathogen [12]. Since the 1980s the old A1 population has been gradually replaced by a new A1/A2 population. This has led to increased virulence and genetic variation worldwide, suggested to be the result of sexual reproduction [13].
Literature Review
Recent years have seen a dramatic intensification in molecular biological studies of P. infestans, including the development of novel tools for transformation and gene silencing and the resources for genetically, transcriptional and physical mapping of the genome [14]. In recent years, there has been a growing movement in the world to reduce the amount of synthetic pesticides being applied to the environment. According to Dhaliwal and Arora, [15] study, only one percent of the total pesticide applied has become effective in controlling pests, remaining 99% goes into various environmental systems. Late blight is one of the most dreaded diseases of potato worldwide and cause significant loss in production. The pathogen is highly variable and adapt to the newly bred varieties and fungicides. Population of P. infestans in most of the countries has changed dramatically and original A1 has almost been displaced by more virulent A2 strain. In India, A2 mating type was recorded in 1990s and now it has displaced the A1 in temperate highlands while in sub-tropical plains still A1 is dominating [16]. Virulence to all major resistance genes has been recorded and in India the racial complexity has reached to its zenith resulting in breakdown of many disease resistant varieties. Indiscriminate use of metalaxyl based fungicides has led to the development of metalaxyl resistance world over including India, which has necessitated the use of additional systemic molecules for the management of this disease [17].
The population of P. infestans characterized using molecular markers has led to better understanding of pathogen at molecular level. Mitochondrial DNA haplotyping of P. infestans has revealed that mt Ia is displacing the other haplotypes globally at a faster rate including India [18].
Disease Cycle and Epidemiology
Disease epidemiology
In the absence of a sexual cycle, the pathogen survives as mycelium in tubers of volunteer plants, seed tubers or discarded tubers near crop fields. Sporangia can also survive several days and even weeks in humid soil; however they do not survive freezing temperatures. Sprouts developed from infected tubers constitute the initial inoculum; mycelium grows through the stem and reaches the soil surface [19]. In a couple of days (4 days in optimal conditions: moderate temperatures and high humidity), after infection has started, new sporangiophores emerge through stomata and produce numerous sporangia that will infect other plants. In just one season, many asexual generations can be produced. Under humid conditions, sporangia located in leaves and stems are washed off and pulled down to the soil where they can produce zoospores and infect tubers near the soil surface. Most of these tubers get infected with rot in the soil through secondary infections produced by other microorganisms, induce infections under inappropriate storage conditions, or mycelium survives on seed tubers until the next season (Figure 1) [19].
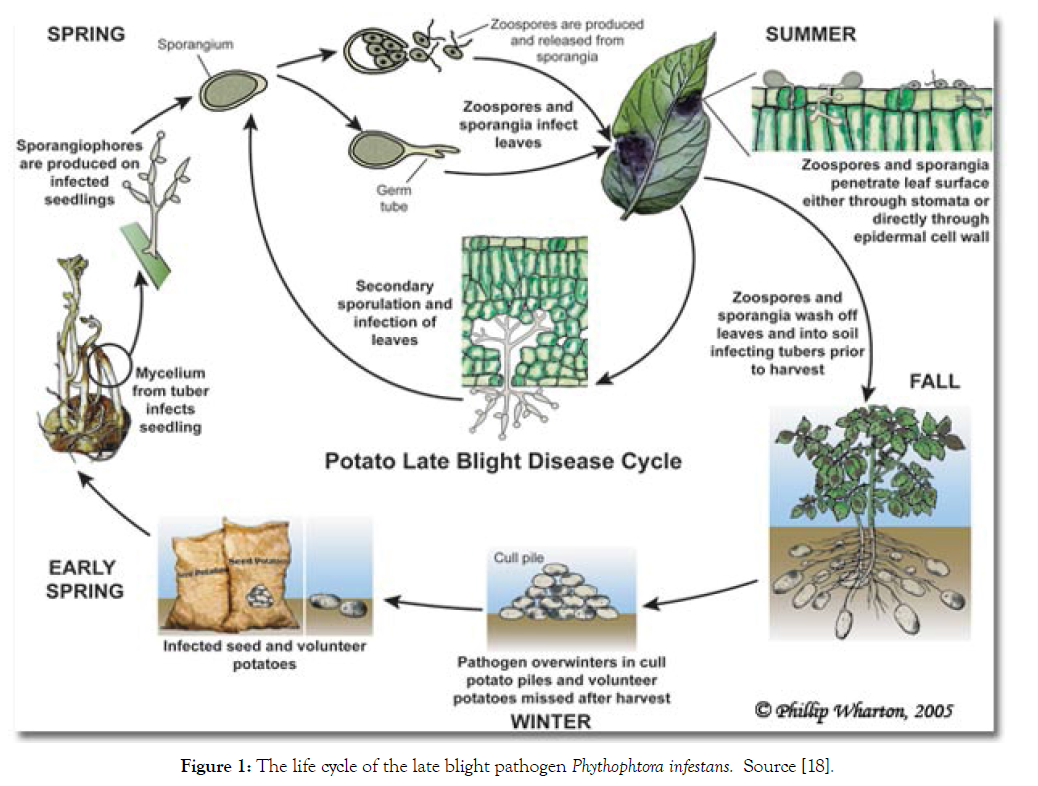
Figure 1: The life cycle of the late blight pathogen Phythophtora infestans. Source [18].
The role of oospores in the epidemiology of potato late blight
Even though coexistence of both mating types is a prerequisite for oospore formation, skewed mating type ratios can result in a large production of oospores [20]. This means that there is a potential for oospore production even in areas where the population is strongly dominated by one mating type. The first observations of oospore formation under field conditions were made in the 1950s in Mexico [21]. In Europe the first reports came 33 or more years later from Germany [22], The Netherlands [23], The United Kingdom [24] and the Nordic countries [25]. These were more sporadic observations of oospore formation.
Favorable conditions for disease development
Excessive humidity (above 90% RH) coupled with suitable temperature for germination of sporangia and further disease development are the principal pre-disposing factors. Where the mean atmospheric temperature exceeds 25°C, the disease is rare or unknown. Cool moist condition favor dispersal or arrival of viable sporangia. Extended dry periods or rapid dehydration can quickly kill many sporangia but within the temperature range of 15°C and 20°C and moisture range of 40 to 88% RH, the life time of sporangia is extended. Maximum spread of the disease occurs when the conditions are favorable for germination of sporangia into zoospores [26].
Yield Loss of Potato by Phytophthora infestans
Potato late blight is considered to be the most serious potato disease worldwide [27]. Yield losses due to the disease are attributed to both premature death of foliage and diseased tubers. In Ethiopia, the disease occurs throughout the major potato production areas and it is difficult to produce the crop during the main rainy season without chemical protection measures [28,29].
In our country Ethiopia and also our continent Africa we used conventional management strategies’ to tackle the potato late blight disease but the disease develops its virulence spectrum due to sexual recombination of the two mating types A1 and A2 mating types.
So our objective will be to familiarize these advanced technologies and review these advanced management strategies.
The main of objectives of this review are
• To understand very well the recent advances on potato late blight disease management up to gene for gene level.
• To incorporate these recent molecular advances with the conventional one for mitigating the virulence spectrum of the pathogen.
The role of molecular plant pathology for potato late blight disease management
Oomycetes are eukaryotic organisms that superficially resemble filamentous fungi, but are phylogenetically related to diatoms and brown algae in the stramenopiles [30-32]. Fossil evidence indicates that a number of oomycetes emerged as endophytes of land plants at least by the Carboniferous period, approximately 300–350 million years ago. Phylogenetic analyses of modern taxa have revealed that plant parasitism has evolved independently in three lineages of oomycetes [33].
Well-known plant pathogens, namely Downy mildews, Phytophthora and Pythium, appear to have radiated from a common plantparasitic ancestor [33]. The impact of oomycetes on humankind is well documented as both a persistent threat to subsistence and commercial farming and as destructive pathogens of native plants [27,34]. As a result, news related to plant diseases caused by oomycetes tends to capture the interest of the general public and is frequently featured in the media. It has produced novel paradigms in understanding host–microbe interactions, effector biology and genome evolution [35,36]. The oomycete community was one of the first in plant pathology to initiate coordinated transcriptome and genome sequencing projects, and subsequently to exploit the resulting resources to drive conceptual advances [37,38]. These days with the genomes serving as unique resources for basic and applied research, oomycetes are best portrayed as a ‘genomicist’s dream’ [38].
Genetic variability
The possible sources of genetic variation in P. infestans are sexual reproduction, mutation, mitotic recombination, para-sexualism, migration, and selection [34]. The markers most used to characterize populations of this pathogen have been virulence, mating type, isozymes, mitochondrial haplotypes, restriction fragment length polymorphism (RFLP) and microsatellites (also known as single sequence repeats or SSR). Furthermore, an increasing number of studies based on sequencing of various nuclear or organelle genes have been developed and the full genomes of a number of isolates have been sequenced. Within the literature related to P. infestans, and a number of other pathogens as well, the term “virulence” has been used as the genetic ability of a P. infestans race (a particular strain) to overcome host resistance, causing a compatibility reaction, that is, the disease occurs [30]. If the R gene product in a plant recognizes the avirulence gene product of a pathogen, rapid death of plant cells near the infection point occurs and the infection is stopped, i.e., there is no disease. Loss or change of avirulence genes leads to what is often called a compatible reaction and disease occurs. The term race groups isolates based on virulence related to R-genes in different potato genotypes. These plants are referred to as “differentials” because they are used to identify the race of a pathogen isolate. Using virulence phenotypes to infer genetic variation in the pathogen population has many constraints because the inference is based on the phenotypic reaction of a pathogen and host without knowing the genetic makeup of either.
Pathogen resistance to fungicides occurs when certain strains have lower sensitivity than normal to a particular product or class of products. This resistance is the result of stable and hereditary mutations. Resistance to the active ingredient metalaxyl and other phenylamides has been reported in P. infestans populations worldwide, becoming a limiting factor when using this type of fungicides. Temporary reduction in sensitivity to a fungicide is an adaptation trait of the pathogen; however, because it is not hereditary, it can be reverted by changing chemical control strategies. Isozymes are variants of an enzyme with the same or similar catalytic activity. Allozymes are a special type of isozymes in which variants are codified by the same locus. Therefore, they are allelic to one another [19].
The P. infestans -potato gene-for-gene interaction
Genetic resistance to P. infestans in both wild and cultivated potato species may be of two forms, either race specific or race nonspecific (field or partial resistance) [39]. Race specific resistance is characterized by interactions between the products of dominant resistance (R) gene alleles in the host and corresponding avirulence (Avr) gene alleles in the pathogen; the so-called gene-for-gene hypothesis [40]. The result is a form of localized programmed cell death called the hypersensitive response (HR) that prevents a further spread of the pathogen. Race nonspecific resistance is poorly under-stood, although recent evidence implies a central role for the HR in all forms of resistance to oomycetes [41].
Genetic studies on P. infestans have been carried out to reveal the genetic basis of avirulence [42]. These studies have revealed that specificity towards R genes in potato is conditioned by single dominant Avr genes for most interactions. Nevertheless, an investigation of different isolates of the pathogen has revealed contradictory results, in that Avr2 and Avr4 were dominant in some isolates and recessive in others. The differences between these strains may be explained either by independent loci determining avirulence in each, or by the occurrence of epistasis inhibitor loci. To facilitate the positional cloning of avirulence genes from P. infestans a genetic linkage map has been constructed using 183 AFLP and 7 RFLP markers [43]. The map contains 10 major linkage groups and represents 1200 cM. Recently, using bulked sergeant analysis, the positions of six dominant Avr loci have been placed on the map [44]. Avr4 was positioned on linkage group (LG) A2- a, Avr2 on LG VI, Avr1 on LG IV and Avr3, Avr10 and Avr11 were shown to be tightly linked on LG VIII. The genome size of P. infestans is 250 Mb, unusually large for an oomycete. A DNA library with large insert sizes and several-fold genome redundancy may thus prove essential for positional cloning [44].
The molecular basis of non-host resistance
Most plant pathogens exhibit specialization and can only infects a limited number of plant species. P. infestans is no exception, primarily infecting the leaves of a variety of Solanaceae. This implies that the majority of non-host plants possess a series of either preformed or inducible mechanisms to successfully prevent infection by this pathogen. Non-host resistance to P. infestans, and indeed to oomycetes in general, involves the HR, presumably activated by the perception of elicitors generated by the pathogen [45]. As with race-specific resistance of potato to P. infestans, non-host resistance may thus involve a gene-for-gene interaction. This is exemplified by the well-characterized interactions between Phytophthora spp. and tobacco. Most Phytophthora spp studied secrete elicitins, 10 kDa holo-proteins that elicit an HR-like response and systemic acquired resistance (SAR) specifically in Nicotiana spp. within the Solanaceae family [46]. Elicitins are thus believed to act as avirulence factors in tobacco–Phytophthora interactions. Similar to many other Phytophthora spp. that has been studied, P. infestans contains a family of elicitin-like genes [47,48]. The gene INF1, encoding the major elicitin secreted by P. infestans, is highly expressed in mycelium grown in various culture media, but is not expressed in sporangia, zoospores, cysts or germinating cysts. During infection, the gene is down-regulated in the early, biotrophic stages of the interaction, but is highly expressed in the later stages of infection, when profuse sporulation and necrosis occur [49]. A gene-silencing strategy has been adopted to inhibit INF1 production. INF1-deficient strains are still pathogenic on potato, but also induce disease lesions when inoculated on Nicotiana benthamiana. In contrast, wild-type P. infestans elicits a typical, localized necrosis on this non-host plant, indicating that INF1 functions as an avirulence factor in the P. infestans–N. benthamiana interaction [50].
The top 10 oomycete pathogens in molecular plant pathology
In the last two decades, increased awareness of the distinctive phylogeny and biology of oomycetes has driven the emergence of a specialist research community that is currently organized under the umbrella of the ‘Oomycete Molecular Genetics Network’. This community has moved the field beyond the gloomy view of the 1980s that oomycetes are a ‘fungal geneticist’s nightmare’ [14,51]. It has produced novel paradigms in understanding host–microbe interactions, effector biology and genome evolution [34,30,51,52].
The Top 10 species and their ranking are: (1) Phytophthora infestans; (2, tied) (2) Hyaloperonospora arabidopsidis;(2, tied) (3) Phytophthora ramorum; (4) Phytophthora sojae; (5) Phytophthora capsici; (6) Plasmopara viticola; (7) Phytophthora cinnamomi; (8, tied) (8) Phytophthora parasitica; (8, tied) (9) Pythium ultimum; and (10) Albugo candida (14). The oomycete community was one of the first in plant pathology to initiate coordinated transcriptome and genome sequencing projects, and subsequently to exploit the resulting resources to drive conceptual advances [51,53]. These days, with the genomes serving as unique resources for basic and applied research, oomycetes are best portrayed as a ‘genomicist’s dream’ [30,51,53].
Tools for the study of gene function in P. infestans
For any aspect of gene function to be assessed in P. infestans, it is essential that a DNA-mediated transformation system is available. The first reported transformation of P. infestans was by Judelson et al. [54] albeit at low efficiency, using protoplasts. Nevertheless, it has been used successfully for anti-sense inhibition of a transgene and for co-transformation using intermolecular ligation [55]. It has also been used to develop an in planta reporter system [50] and for heterologous expression in P. infestans of a gene from another Phytophthora spp. [56]. However, the transformation procedure used in these reports required the digestion of the cell wall using Novozym 234, an enzyme mixture that is no longer produced. Therefore, different methods for transformation must be adopted. One such method, utilizing microprojectile bombardment to transform P. infestans, has recently been reported [14,52] and overcomes the requirement for protoplast formation with Novozym. For map-based cloning of genes and studies of genome structure and organization, two BAC libraries have been constructed from P. infestans. Randall and Judelson [57] reported the generation of a library comprising four folds genome coverage and an average insert size of 75 kb. Birch et al. [58] have generated a BAC library comprising 10-fold genome coverage and an average insert size of 98 kb. The transformation of entire BAC clones into P. infestans, making it possible to determine the presence of a gene, such as an avirulence gene, within large regions of the genome [57]. P. infestans is diploid, making it difficult to perform gene knockouts, as is often done in the fungi that have a haploid stage in their life cycle. The occurrence of post-transcriptional gene silencing in P. infestans is therefore a promising development in the area of P. infestans functional genomics [59]. However, at this time gene silencing in P. infestans is not yet routine and its molecular basis is poorly understood. The utility of gene silencing as a functional tool in P. infestans has been shown by the development of transgenic strains silenced for transcription of the elicitin encoding inf 1 gene [50]. Functional analysis of cloned P. infestans genes may also be performed without transformation or silencing in P. infestans.
The P. infestans –plant interaction transcriptome
A number of developmental processes are required for P. infestans to successfully invade its plant host, including the formation of zoospores, their encystment, the production of a germ tube, and the development of appressoria, hyphae, haustoria and, finally, sporangiophores [58]. Successful infection and the development of disease symptoms are often termed a compatible interaction. If the plant, through mounting a series of defenses, is able to interrupt or inhibit any of these processes, colonization of the pathogen, and its further spread to other plants, will be prevented. This is often called an incompatible interaction. The perception of P. infestans by its host, and the ability of the pathogen to avoid or overcome the host’s defenses, implies a complex, dynamic communication network between the interacting organisms. The induction of biochemical response pathways, or the development of cell type specific to the interaction, requires the up- or down-regulation of countless genes. We have recently coined the term ‘interaction transcriptome’ to mean the sum of the transcripts, from both host and pathogen that are produced during their association [59]. A key initial step in understanding the mechanisms and processes involved in the P. infestans–plant interaction involves determining the interaction transcriptome.
The development of low-cost, high throughput DNA sequencing has allowed plant pathology to enter the ‘genomics era’. In particular, projects involving large scale sequencing of cDNAs (Expressed Sequence Tags or ESTs) are on-going for a wide variety of crop plants. EST information has also emerged from P. infestans [48] and P. sojae [60]. Approximately 2000 – 3000 ESTs from each species are currently housed in the Phytophthora Genome Initiative (PGI) data-base [61].
Thus, in silico analyses can prove powerful in distinguishing candidate plant and pathogen genes in the Phytophthora –plant interaction transcriptome, although it remains to be seen whether clear differences in GC content between host and pathogen cDNAs will be observed in other patho-systems. Many proteins that play a role in pathogenicity, or elicit a defense response in the plant, are likely to be surface components of the pathogen. Tyler et al. [62] have been exploiting the increasing EST data to search for genes encoding potential extracellular proteins in P. infestans. They have developed an algorithm called PEXFINDER V1.0 (where PEX represents Phytophthora extracellular protein) to rapidly identify putative secreted or membrane-associated proteins encoded by the ESTs, with the expectation that candidate genes with an essential survival or virulence role would be targeted for downstream functional analyses. To date, analysis of more than 2000 ESTs has revealed 145 independent Pex genes, of which 85 show no similarity to sequences in international databases. Many of the genes with a key role in either host defense or pathogenicity will be up-regulated during the P. infestans–plant association. A number of methods exist for isolating such differentially expressed genes. [63] Judelson et al. [55] isolated a number of in planta induced (ipi ) genes by differential screening of a P. infestans genomic DNA library with cDNA derived from mRNA prepared after infection of potato leaves with P. infestans and from mRNA prepared after growth of P. infestans on a basic medium in culture. Amongst the up-regulated P. infestans sequences were the ipiO and ipiB genes, each of which is a member of a cluster of related sequences [63]. The ipiO gene has since been shown to be expressed in invading hyphae during the early stages of infection [59].
Using standard subtractive hybridization techniques, Gornhaldt et al. [64] isolated a family of mucin-like genes, termed car genes that are up-regulated in germinating cysts shortly before the onset of infection. The car genes were shown to be clustered in the genome. The authors postulated that the car gene products may serve to provide a mucous cover to protect the germinating cysts from desiccation, physical damage and plant defenses.
More recently, a PCR-based method for isolating differentially expressed genes, suppression subtractive hybridization (SSH), has been used to study potato –P. infestans interactions. This technique can be readily combined with large-scale sequencing approaches and allows the detection of low-abundance differentially expressed transcripts. This is a major advantage in analyzing plant–microorganism associations, where often only small amounts of biological material are available. SSH has been used to isolate potato genes that are up-regulated in the incompatible and compatible interactions with P. infestans [65,66]. Moreover, SSH-derived cDNA populations enriched for sequences expressed specifically at early (15 h post inoculation) and late (72 h post-inoculation) stages of infection have been generated and used as probes to screen the P. infestans BAC library constructed by Birch and Kamoun [58]. Each probe hybridized to a number of BACs, but neither hybridized to the same clones. Quantitative RT-PCR has been used to demonstrate that these BACs contain sequences that are upregulated specifically at the early or late stages of infection and are thus candidates for a role in pathogenicity [67].
Management of Potato Late Blight
Cultural control
There are different methods of cultural practices applicable for late blight management. Cultural practices are the first line of defense against late blight [68,69]. Late blight is controlled by eliminating cull piles and volunteer potatoes, using proper harvesting and storage practices, and applying fungicides when necessary [70]. The most effective strategy for managing late blight is to avoid sources of inoculum. The initial sources of inoculum are likely to be infected potatoes in cull piles, infected volunteer potato plants that have survived the winter, and infected seed tubers. Therefore, it is important to keep a clean operation by destroying all cull and volunteer potatoes [27].
Chemical control
Fungicides that are used against late blight can be classified into two basic mobility groups: protectant or penetrant. Fungicides can slow or stop the development of new symptoms if applied in a timely fashion, but fungicides will not cure existing light blight symptoms [71]. In Ethiopia the first spray with Ridomil MZ 63.5% WP at a rate of 2 kg ha-1 followed by 2-3 sprays (need base application) of Dithane M-45 (Mancozeb) at a rate of 3 kg ha-1 were found to be effective in controlling late blight [72,73]. He also reported that, reduced rates of Ridomil application resulted in better management of potato late blight with the highest marginal rate of return.
Anti-resistance management strategies
• Restrict the number of high-risk fungicide applications. Mix a high-risk fungicide with a low-risk fungicide to be sure spores will not survive.
• Alternate applications of high-risk fungicides with low-risk fungicides, including the use of fungicides with different modes of action. Add other integrated management practices, different from those of the chemical component in order to avoid disease development.
Biological control
Biological control of crop disease is receiving increased attention as an environmentally sound alternative to chemical pesticides. Some species of Trichoderma and Pseudomonas are among the major microorganisms that have shown great potential for biological control of several plant pathogens. Trichoderma species have shown bio control potential against many plant pathogens including diseases caused by Sclerotinia minor [74,75]. Botanical control is one of the safe substitutes to be explored to control this phyto-pathogen. In the present study an attempt has been made to evaluate the antifungal activity of plant extracts against the above pathogen.
Crude ethanolic extracts of five different plant materials viz. Brassica nigra, Cinnamomum camphora, Eupatorium adenophorum, Lantana camara and Melia azedarach were screened and tested against the fungal isolate of P. infestans [76]. The antifungal activity of the crude extracts was evaluated by agar well diffusion method and two fold broth dilution method. The moisture content was highest in the twigs of L. camara and lowest in the cake of B. nigra. C. camphora gave the highest yield of 70% while M. azedarach had the lowest yield of (9.75%) of crude extracts. B. nigra was found most effective against P. infestans with both MIC and MFC values at 6.25 mg/ml while C. camphora was found least effective [76]. Different types of plant extracts with different concentration significantly (P= <0.05) inhibited the growth of the pathogen. The extracts of B. nigra, E. adenophorum, L. camara and M. azedarach inhibited the growth of P. infestans from 10 mg/ml concentration, B. nigra being the most effective one with both MIC and MFC values 6.25 mg/ml. C. camphora showed the inhibitory activity only from the 30 mg/ml concentration [76].
Host-plant resistance
Host resistance to late blight is of significance in integrated late blight management due to its long-term economic benefits for farmers. It also minimizes changes in the population structure of P. infestans, decreasing the likelihood of fungicide resistance [77,78]. The use of resistant varieties is among the most effective and environmentally safe means of managing the disease. Biotechnology is also being employed in the pursuit of late blight resistance. Genetically-engineered plants, however, are not acceptable for organic production [79]. But, potato cultivars with high blight resistance can be destroyed by new strains of the fungus since the resistance is controlled by single gene. The use of durable or polygenic resistance is sometimes interpreted to be synonymous with intermediate resistance levels but cultivars ranging from complete susceptibility to very high resistance. Polygenic resistance has proved to be helpful in reducing the amount of fungicides [74]. Use of resistant varieties is one of the main components of late blight management and is especially effective under tropical conditions [80]. However, the race-specific Oligo-genic resistance in the existing released potato varieties can be rapidly broken down by compatible races of P. infestans rendering the varieties to be susceptible to the disease within a short period [80,81].
Generally resistant potato varieties and improved cultural practices can reduce late blight [1]. It is not sufficient to rely on varietal resistance to control late blight, as, in favorable weather, late blight can severely affect these varieties unless they are sprayed with a good protective fungicide. Even resistant varieties should be sprayed regularly with fungicides to eliminate, as much as possible, the possibility of becoming suddenly attacked by races of the fungus to which they are not resistant. According to Binyam et al. [72] onset of the potato late blight disease was delayed almost by 20 days on the moderately resistant varieties as compared to the moderately susceptible and susceptible varieties.
Genetic control
Genetic control refers to the use of varieties or species of the host that have resistance to the pathogen which acts to stop or slow down disease development. There are two ways that resistance to P. infestans is expressed in the potato plant. The first one is characterized by triggering a hypersensitivity response (HR) as small necrotic lesions and is called race-specific resistance, vertical resistance, qualitative resistance, unstable resistance or complete resistance. It is governed by R genes with strong effects that produce products which in turn interact with products of avirulence genes (Avr) of the pathogen. Most major genes known until now mainly come from S. demissum, however a number of new genes have recently been detected in S. bulbocastanum and other Solanum spp. This resistance is race-specific; its inheritance is qualitative and in the past never had long duration. The exact way in which products of R genes and Avr genes interact is unknown; however, diverse models have been proposed.
The second type of resistance is governed by minor genes of additive effect and is called general resistance, quantitative resistance, and polygenic resistance, non-specific resistance, and partial resistance, horizontal or field resistance. Inheritance is quantitative and because it is governed by several or many genes, it is theoretically more stable and effective against all the pathogen races. Furthermore, adaptation for greater pathogen aggressiveness in host genotypes with general resistance has been identified. Integrating genetic resistance and chemical control helps, in reducing the use of fungicides, decreases production costs and reduces damage to human health and the environment [82].
Integrated Disease Management (IDM)
Effective management of this disease requires implementation of an integrated disease management approach. Although the most important measures are cultural, resistant cultivars and chemical controls should also be utilized. Integrated pest management has helped farmers drastically reduce the need for chemical controls while increasing production [1]. Effective control of late blight requires implementing an integrated disease management approach [68,69,83]. Integration of different management options, including cultural practices (good crop husbandry), resistant varieties and fungicides is required to control late blight. The integration of reduced rate of Ridomil application and moderately resistant potato varieties, in the management of potato late blight is very important in reducing environmental pollution and input cost of the fungicide, and increase in production and profitability of high quality potato tuber yield [72]. For effective control of late blight, integrated management must be adopted by all producers, including large and small-scale farmers. Experimental plots with IDM-LB yielded 50% and 75% more than late planting (planting during the month recommended for potato-growing) alone [84]. According to Binyam et al. [72] cost effective management of late blight was obtained by integrating potato varieties with the lowest rate of Ridomil application. The combinations of the varieties Gabissa, Chiro, Harchassa, Bedassa and Zemen with 0.75 kg ha-1 Ridomil application were resulted in up to 28, 21, 18, 16 and 13% marginal rate of returns, respectively [73].
Summary and Conclusion
Late blight of potato is the most dreaded disease and will continue to remains as the pathogen is evolving at a fast rate and adapting to new environments and hosts. There is need to characterize the pathogen population with more robust molecular markers and to study the epidemiology of isolates grouped in different categories on the basis of markers. Disease resistant varieties should be developed mode of action need to be identified and used along with compatible bio-control agents to minimize the use of pesticides. As more and more information is being generated there is a need to develop an appropriate disease management strategy based on farmer friendly information technology.
Future Directions
Molecular works must be done intensively for the most important crops such as potato, wheat, coffee, ground-nut, sesame, cotton etc. which have multiple benefit for human being used as food, as income generating item. It gives high yield with in small plot of land around garden and exportable crops as foreign currency for Ethiopia. Pathogen will be detected at molecular level using marker assisted selection. Highly skilled man power must be trained to detect the pathogen and give remedies for the problem at its infant stage.
Acknowledgments
First of all, we would like to thank the Almighty God and St. Mary for making all things possible with their boundless and kind supply of unconditional supports. We have also appreciated the way of coordination and interaction to publish this paper.
REFERENCES
- FAO (Food and Agriculture Organization). Potato World: Africa-International Year of the Potato 2008.
- Chandrakala A, Chandrashekar SC, Jyothi G, Ravikumar BM. Effect of cell-free culture filtrates of bio-control agents on the spore germination and infection by Phytophthora infestans causing late blight of potato. Glob J Biol Agricult Health Sci. 2012;1(2):40-45.
- Byrne PF, Volk GM, Gardner C, Gore MA, Simon PW, Smith S. Sustaining the future of plant breeding: The critical role of the USDA‐ARS National Plant Germplasm System. Crop Sci. 2018;58(2):451-468.
- Crowell EF, McGrath JM, Douches DS. Accumulation of vitamin E in potato (Solanum tuberosum) tubers. Transgenic Res. 2008;17(2):205-217.
- CSA (Central Statistical Authority). Report on farm management practices. (Private peasant holdings) Addis Ababa, Ethiopia. 2012.
- Mekonnen D. Effects of integrated nutrient management on Agronomic performance of potato (Solanum tuberosum L.) and fertility of nitosol at Bako. Haramaya University M.Sc. Thesis. 2006.
- Fry WE, Goodwin SB. Resurgence of the Irish potato famine fungus. Bioscience. 1997;47(6):363-371.
- Adolf B, Andrade-Piedra J, Molina FB, Przetakiewicz J, Hausladen H, Kromann P, et al. Fungal, Oomycete, and Plasmodiophorid Diseases of Potato. InThe Potato Crop. 2020;307-350.
- Berkeley RM, De Bary A. Potato Late Blight. Molecular Tools to Unravel the Role of Genes fromPhytophthora infestans. 2010;1.
- Arora RK, Sharma S, Singh BP. Late blight disease of potato and its management. Potato Journal. 2014;41(1).
- Haverkort AJ, Struik PC, Visser RG, Jacobsen EJ. Applied biotechnology to combat late blight in potato caused by Phytophthora infestans. Potato Res. 2009;52(3):249-264.
- Goodwin SB, Cohen BA, Deahl KL, Fry WE. Migration from northern Mexico as the probable cause of recent genetic changes in populations of Phytophthora infestans. Phytopathology. 1994;84:553-558.
- Fry WE, Goodwin SB, Dyer AT, Matuszak JM, Drenth A, Tooley PW, et al. Historical and recent migrations of Phytophthora infestans : chronology, pathways and implications. Plant Dis. 1993:653-661.
- Kamoun S, Furzer O, Jones JD, Judelson HS, Ali GS, Dalio RJ, et al. The Top 10 oomycete pathogens in molecular plant pathology. Molecular plant pathology. 2015;16(4):413-434.
- Dhaliwal GS, Arora R. Integrated pest management concepts and approaches. Kalyani Publishers, New Delhi, India. 2001:66-67.
- Fry WE, Birch PR, Judelson HS, Grünwald NJ, Danies G, Everts KL, et al. Five reasons to consider Phytophthora infestans a reemerging pathogen. Phytopathology. 2015;105(7):966-981.
- Chowdappa P, Kumar NB, Madhura S, Kumar MS, Myers KL, Fry WE, et al. Emergence of 13_ A 2 Blue Lineage of Phytophthora infestans was Responsible for Severe Outbreaks of Late Blight on Tomato in South‐West India. J Phytopathol. 2013;161(1):49-58.
- Wharton PS. Potato disease in Michigan. Crop and Soil Sciences Extension Bulletin. 2005:2945.
- Thomas‐Sharma S, Abdurahman A, Ali S, Andrade‐Piedra JL, Bao S, Charkowski AO, et al. Seed degeneration in potato: the need for an integrated seed health strategy to mitigate the problem in developing countries. Plant Pathol. 2016;65(1):3-16.
- Cohen Y, Farkash S, Reshit Z, Baider A. Oospore production of Phytophthora infestans in potato and tomato leaves. Phytopathology. 1997;87(2):191-196.
- Fry WE. Phytophthora infestans: The itinerant invader;“late blight”: the persistent disease. Phytoparasitica. 2020;48(1):87-94.
- Yuen JE, Andersson B. What is the evidence for sexual reproduction of P hytophthora infestans in E urope?. Plant Pathol. 2013;62(3):485-491.
- Drenth A, Turkensteen LJ, Govers F. The occurrence of the A2 mating type of Phytophthora infestans in the Netherlands; significance and consequences. Neth J Plant Pathol. 1993;99(3):57-67.
- Hanson K, Shattock RC. Formation of oospores of Phytophthora infestans in cultivars of potato with different levels of race‐nonspecific resistance. Plant Pathol. 1998;47(2):123-129.
- Andersson B, Sandström M, Strömberg A. Indications of soil borne inoculum ofPhytophthora infestans. Potato Res. 1998;41(4):305-310.
- Legard DE, Lee TY, Fry WE. 2010. Pathogenic specialization in Phytophthora infestans: Agressiveness on tomato. Phytopathology. 2010;85:1362.
- Agrios GN. Plant Pathol. (5th Edn). Academic Press, London, 2005:922.
- Borgal HB, Arend C, Jacobi, Kanyarukis S, Kulazia A, Lemaga BL, et al. Production, marketing and consumption of potato in the Ethiopia highlands’ (Holleta, Awassa, and Alemaya) Center of Advanced training in agricultural development technology. University of Berlin. 1980.
- Kasa B, Woldegiorgis G. Effect of planting dates on late blight severity and tuber yields of different potato varieties. Pest Management Journal of Ethiopia. 2000;4:51-63.
- Jiang RH, Tyler BM. Mechanisms and evolution of virulence in oomycetes. Annu Rev Phytopathol. 2012;50:295-318.
- Yoshida K, Burbano HA, Krause J, Thines M, Weigel D, Kamoun S. Mining herbaria for plant pathogen genomes: back to the future. PLoS Pathog. 2014;10(4):e1004028.
- Demissie YT. Integrated potato (Solanum tuberosum L.) late blight (Phytophthora infestans) disease management in Ethiopia. AJBIO. 2019;7(6):123-130.
- Yoshida K, Schuenemann VJ, Cano LM, Pais M, Mishra B, Sharma R, et al. The rise and fall of the Phytophthora infestans lineage that triggered the Irish potato famine. Elife. 2013;2:e00731.
- Haas BJ, Kamoun S, Zody MC, Jiang RH, Handsaker RE, Cano LM, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461(7262):393-398.
- Vleeshouwers VG, Raffaele S, Vossen JH, Champouret N, Oliva R, Segretin ME. Understanding and exploiting late blight resistance in the age of effectors. Annu Rev Phytopathol. 2011;49:507-531.
- Zhang F, Chen H, Zhang X, Gao C, Huang J, Lü L, et al. Genome analysis of two newly emerged potato late blight isolates sheds light on pathogen adaptation and provides tools for disease management. Phytopathology®. 2021;111(1):96-107.
- Mahadevakumar S, Sridhar KR. Plant-Microbe Interaction: Current Developments and Future Challenges. InAdvances in Plant Microbiome and Sustainable Agriculture. 2020:1-38.
- Ochola S, Huang J, Ali H, Shu H, Shen D, Qiu M, et al. Editing of an effector gene promoter sequence impacts plant‐Phytophthora interaction. J Integr Plant Biol. 2020;62(3):378-392.
- Wastie RL. Resistance to powdery scab of seedling progenies of Solanum tuberosum. Potato Res. 1991;34(3):249-352.
- Flor HH. Current status of the gene-for-gene concept. Annu Rev Phytopathol. 1971;9(1):275-296.
- Nair RA, Kiran AG, Sivakumar KC, Thomas G. Molecular characterization of an oomycete-responsive PR-5 protein gene from Zingiber zerumbet. Plant Mol Biol Rep. 2010;28(1):128.
- Waller JM, Cannon PF. Fungi as plant pathogens. Plant Pathologist’s Pocketbook 3rd Edition. 2002:85.
- Van der Lee T, De Witte I, Drenth A, Alfonso C, Govers F. AFLP linkage map of the oomycetePhytophthora infestans. Fungal Genet Biol. 1997;21(3):278-291.
- Van der Lee T, Robold A, Testa A, Van’t Klooster JW, Govers F. Mapping of avirulence genes in Phytophthora infestans with amplified fragment length polymorphism markers selected by bulked segregant analysis. Genetics. 2001;157(3):949-956.
- Kamoun S, Huitema E, Vleeshouwers VG. Resistance to oomycetes: a general role for the hypersensitive response?. Trends in Plant sci. 1999;4(5):196-200.
- Paris R, Lamattina L. Phytophthora infestans secretes extracellular proteases with necrosis inducing activity on potato. European journal of plant pathology. 1999;105(8):753-760.
- Kamoun S, Lindqvist H, Govers F. A novel class of elicitin-like genes from Phytophthora infestans. Mol Plant-Microbe Interact. 1997;10(8):1028-1030.
- Kamoun S, Hraber P, Sobral B, Nuss D, Govers F. Initial assessment of gene diversity for the oomycete pathogen Phytophthora infestans based on expressed sequences. Fungal Genet Biol. 1999;28(2):94-106.
- Kamoun S, Van West P, De Jong AJ, De Groot KE, Vleeshouwers VG, Govers F. A gene encoding a protein elicitor of Phytophthora infestans is down-regulated during infection of potato. Molecular Plant-Microbe Interactions. 1997;10(1):13-20.
- Kamoun S, Van West P, Govers F. Quantification of late blight resistance of potato using transgenic Phytophthora infestans expressing β-glucuronidase. Eur J Plant Pathol. 1998;104(5):521-525.
- Schornack S, Huitema E, Cano LM, Bozkurt TO, Oliva R, Van Damme M, et al. Ten things to know about oomycete effectors. Mol Plant Pathol. 2009;10(6):795-803.
- Dean R, Van Kan JA, Pretorius ZA, Hammond‐Kosack KE, Di Pietro A, Spanu PD, et al. The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. 2012;13(4):414-430.
- Pais M, Win J, Yoshida K, Etherington GJ, Cano LM, Raffaele S, et al. From pathogen genomes to host plant processes: the power of plant parasitic oomycetes. Genome Biol. 2013;14(6):1-10.
- Judelson HS, Tyler BM, Michelmore RW. Transformation of the oomycete pathogen, Phytophthora infestans. Mol Plant-Microbe Interact. 1991;4:602-607.
- Judelson HS, Dudler R, Pieterse CJ, Unkles SE, Michelmore RW. Expression and antisense inhibition of transgenes in Phytophthora infestons is modulated by choice of promoter and position effects. Gene. 1993;133(1):63-69.
- Panabieres F, Birch PR, Unkles SE, Ponchet M, Lacourt I, Venard P, et al. PLANT-MICROBE INTERACTIONS-Heterologous expression of a basic elicitin from Phytophthora cryptogea in Phytophthora infestans increases its ability to cause leaf necrosis in tobacco. Microbiology-Reading. 1998;144(12):3343-3350.
- Randall TA, Judelson HS. Construction of a bacterial artificial chromosome library of Phytophthora infestans and transformation of clones into P. infestans. Fungal Genet Biol. 1999;28(3):160-170.
- Birch PR, Kamoun S. Studying interaction transcriptomes: coordinated analyses of gene expression during plant-microorganism interactions. New technologies for life sciences: a trends guide. 2000;1931:77-82.
- Van West P, Kamoun S, van’t Klooster JW, Govers F. Internuclear gene silencing in Phytophthora infestans. Molecular cell. 1999;3(3):339-348.
- Qutob D, Hraber PT, Sobral BW, Gijzen M. Comparative analysis of expressed sequences in Phytophthora sojae. Plant Physiol. 2000;123(1):243-254.
- Lengeler KB, Davidson RC, D'souza C, Harashima T, Shen WC, Wang P, et al. Signal transduction cascades regulating fungal development and virulence. MMBR. 2000;64(4):746-785.
- Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, Aerts A, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313(5791):1261-1266.
- Pieterse CJ, Van West P, Verbakel HM, Brassé PW, Van den Berg-Velthuis GC, Govers F. Structure and genomic organization of the ipiB and ipiO gene clusters of Phytophthora infestans. Gene. 1994;138(1-2):67-77.
- Görnhardt B, Rouhara I, Schmelzer E. Cyst germination proteins of the potato pathogen Phytophthora infestans share homology with human mucins. Mol Plant-Microbe Interact. 2000;13(1):32-42.
- Avrova AO, Stewart HE, De Jong W, Heilbronn J, Lyon GD, Birch PR. A cysteine protease gene is expressed early in resistant potato interactions with Phytophthora infestans. Mol Plant-Microbe Interact. 1999;12(12):1114-1119.
- Dellagi A, Heilbronn J, Avrova AO, Montesano M, Palva ET, Stewart HE, et al. A potato gene encoding a WRKY-like transcription factor is induced in interactions with Erwinia carotovora subsp. atroseptica and Phytophthora infestans and is coregulated with class I endochitinase expression. Molecular plant-microbe interactions. 2000;13(10):1092-1101.
- Whisson SC, Avrova AO, Boevink PC, Armstrong MR, Seman ZA, Hein I, et al. Exploiting knowledge of pathogen effectors to enhance late blight resistance in potato. Potato Res. 2011;54(4):325-340.
- Kirk W. Potato late blight alert for the Midwest. Field Crop Advisory Team Alert Curent News Articles. 2009.
- Kirk W, Wharton P, Hammerschmidt R, Abu-El Samen F, Douches D. Late Blight. Michigan State University Extension Bulletin. 2013:2945.
- Davis RM, Nunez J, Aegerter BJ. Potato Late Blight. Statewide IPM Program, Agriculture and Natural Resources, University of California, USA. 2009.
- Beckerman J. Understanding fungicide mobility. Purdue Extension BP-70-W. 2008.
- Tsedaley B, Hussen T, Tsegaw T. Efficacy of reduced dose of fungicide sprays in the management of late blight (Phytophthora infestans) disease on selected potato (Solanum tuberosum L.) varieties Haramaya, Eastern Ethiopia. Int J Agric Biol Healthc. 2014;4(20):46-52.
- Tsedaley B, Hussen T, Tsegaw T. Tuber yield loss assessment of potato (Solanum tuberosum L.) varieties due to late blight (Phytophthora infestans) and its management Haramaya, Eastern Ethiopia. J Biol Agric Healthc. 2014;4:45-54.
- Jones EE, Stewart A. Biological control of Sclerotinia minor in lettuce using Trichoderma species. InProceedings of the New Zealand Plant Protection Conference 1997;50:154-158.
- Dolatabadi KH, Goltapeh EM, Varma A, Rohani N. In vitro evaluation of arbuscular mycorrhizal-like fungi and Trichoderma species against soil borne pathogens. J Agric Technol. 2011;7(1):73-84.
- Messgo-Moumene S, Boukhalfa R, Belaïdi D. In vitro antifungal activity of different plant extracts against Phytophthora infestans the causal agent of potato late blight. New European Union plant health regime: A more stringent regulation that could impact trade from developing countries in the near future. 2017;12(1).
- Hakiza JJ. The importance of resistance to late blight in potato breeding in Africa. InProceedings of the Global Initiative on Late Blight Conference, Quito, Ecuador. 1999:16-19.
- Mukalazi J, Adipala E, Sengooba T, Hakiza JJ, Olanya M, Kidanemariam HM. Metalaxyl resistance, mating type and pathogenicity of Phytophthora infestans in Uganda. J Crop Prot. 2001;20(5):379-388.
- Shapiro L, Hager M. Is organic better?. Newsweek. 1998;131(22):54-57.
- Shtienberg D, Raposo R, Bergeron SN, Legard DE, Dyer AT. Incorporation of cultivar resistance in a reduced-sprays strategy to suppress early and late blights on potato. Plant Dis. 1994;78(1):23-26.
- CIP (International Potato Center). Fungal diseases of potato. Report of planning conference on fungal diseases of the potato. CIP.1989:216.
- Pérez W, Forbes G. Potato late blight: Technical manual [Chinese]. 2010.
- Martin AD, Neil CG, Arthur HL, Duane P. Leaf blight diseases of potato. North Dakota State University Agriculture and University Extension. 1994.
- Goutam U, Thakur K, Salaria N, Kukreja S. Recent Approaches for Late Blight Disease Management of Potato Caused by Phytophthora infestans. InFungi and their Role in Sustainable Development: Current Perspectives 2018:311-325.
Citation: Tadesse Y, Kesho A, Amare D (2021) Recent Advances in Potato late Blight Disease Management Strategies. J Plant Pathol Microbiol 12:559.
Copyright: © 2021 Tadesse Y, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

