Indexed In
- Academic Journals Database
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- China National Knowledge Infrastructure (CNKI)
- Scimago
- Ulrich's Periodicals Directory
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- MIAR
- University Grants Commission
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Short Communication - (2021) Volume 0, Issue 0
Rapid Screening for Variants of Concern in Routine SARS-CoV-2 PCR Diagnostics
Paul Naaber1,2*, Andrio Lahesaare1, Laura Truu1, Andres Soojärv1, Ainika Adamson1, Kaido Beljaev1, Rainar Aamisepp1 and Kaspar Ratnik12Department of Microbiology, Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia
Received: 06-May-2021 Published: 28-May-2021, DOI: 10.35248/2157-7560.21.s13.005
Description
The emerging spread of Variants of Concern (VOC) of SARSCoV- 2 has been noted in several countries worldwide during the few last months. Such variants (e.g. B.1.1.7, B1.351, and P.1) are associated with increased transmissibility and morality, and possibly also the reduction of vaccine effectiveness [1-4]. Realtime surveillance with a prompt response is essential for the containment of their future spread.
Sequencing of positive samples is the gold standard for the typing classification of SARS-CoV-2 strains and also the identification of VOCs. However, this method is expensive and time-consuming. Although many countries have made a great effort to increase sequencing capacity it still only covers a minority of cases and the results only become available after several days or weeks. For prompt actions, largescale surveillance with cheaper and rapid methods is required uniformly.
Recently several RT-PCR tests for the detection of SARS-CoV-2 mutations have been developed and this approach is also recommended by ECDC [5-7]. The detection of S-gene drop-out by some SARSCoV-2 assays has also been shown as a useful tool for screening B.1.1.7.
For rapid screening of VOC, we introduced S-dropout routine monitoring in combination with VOC screening by RT-PCR in SYNLAB Estonia, Tallinn. SYNLAB is serving all of the Estonian regions and performing 79% of all Estonian SARSCoV- 2 PCR tests, so representing the whole country’s situation.
We use Taq Path COVID-19 CE-IVD RT-PCR (Thermo Fisher Scientific Inc.) for routine SARS-CoV-2 testing which is able to detect S-dropout associated with Del 69-70. S-dropouts’ counts and proportion from positive results by counties and patient groups together with trend analysis is updated daily on the COVID-19 diagnostics dashboard (Figures 1-4).
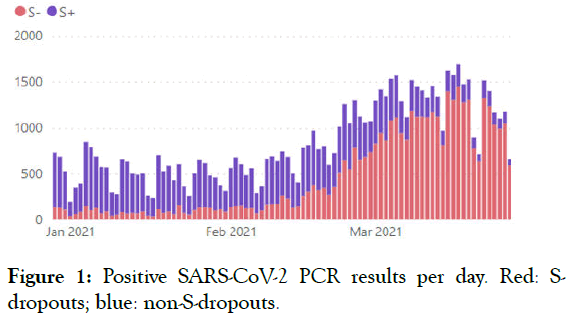
Figure 1: Positive SARS-CoV-2 PCR results per day. Red: Sdropouts; blue: non-S-dropouts.
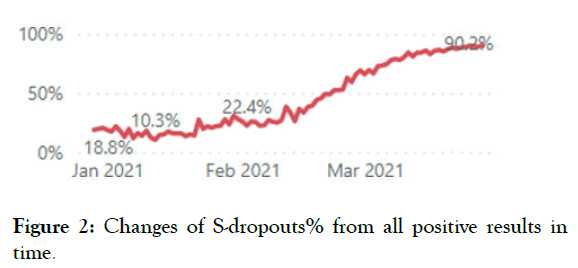
Figure 2: Changes of S-dropouts% from all positive results in time.
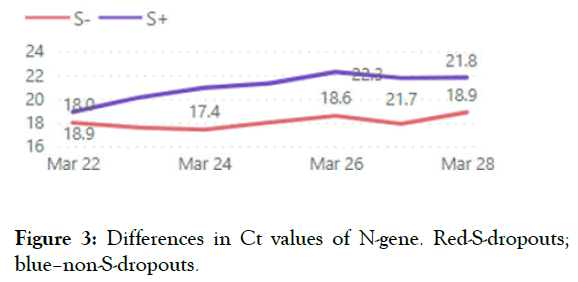
Figure 3: Differences in Ct values of N-gene. Red-S-dropouts; blue–non-S-dropouts.
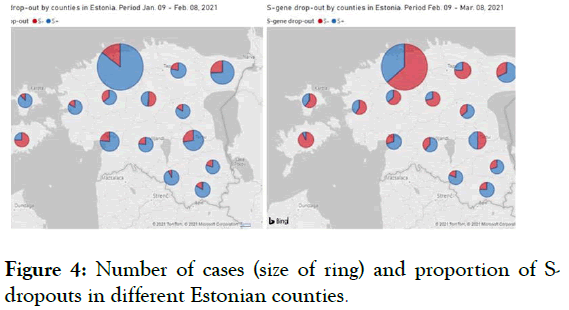
Figure 4: Number of cases (size of ring) and proportion of Sdropouts in different Estonian counties.
As a pilot study, we screened 1116 positive results (a random selection from a total drawn from 1229 positive results of March 08, 2021) for Del 69-70; N501Y ja E484K by Novaplex SARSCoV- 2 Variant Assay (Seegene Inc.) to evaluate the country’s situation at the moment.
The S-dropout proportion has varied from 20% to 96% (average 78%) of all positive samples. From all of the reviewed Sdropouts, 95% were of the UK variant B.1.1.7. Thus, B.1.1.7 appeared to be the predominant genotype in Estonia, causing a total of 74% of all COVID-19 cases on this testing day.
Two positive samples (0.18%) carried N501Y and E484K mutations indicating to B1.351 or P.1 variant.
For samples with N501Y and E484K mutation and randomly selected other strains amounting to 134 the sequencing was performed in the SYNLAB MVZ Humangenetik Mannheim GmbH (Germany) using commercially available Illumina COVID Seq Test (Illumina Inc.). All UK variants (B.1.1.7) detected by RT-PCR were also confirmed by sequencing and S-dropouts without N501Y mutations belonged to lineage B.
1.258. The same strain B.1.258 has been already described in the Czech Republic and Slovakia in the end of 2020 [8]. Two samples with N501Y and E484K mutations were confirmed as B.1.351 (South African variant).
Conclusion
Continuous monitoring of S-dropouts is a useful and inexpensive tool for a follow-up of the UK variants epidemiology. However, regular cross-sectional studies for the screening of all relevant VOC should be performed since the proportion of UK strains in S-drop-outs is not stable and has significantly increased over the last 3 months in Estonia according to our data. Moreover, other important VOCs can appear and the UK strain can acquire additional mutations such as E484K.
The rapid detection of these variants means paying special attention to particular regions and patient groups as well, which should be mandatory in border SARS-CoV-2 screening, followed by prompt actions for containment of these variants.
In conclusion, RT-PCR detection of particular mutations is generally an easy and reliable way to screen thousands of samples during a short time and, together with S-dropout automated monitoring, is a useful tool for VOC containment. This approach can significantly support regular sequencingbased surveillance.
REFERENCES
- Plante JA, Mitchell BM, Plante KS, Debbink K, Weaver SC, Menachery VD. The variant Gambit: COVID’s next move. Cell Host & Microbe. 2021.
- Di Caro A, Cunha F, Petrosillo N, Beeching NJ, Ergonul O, Petersen E, et al. SARS-CoV-2 escape mutants and protective immunity from natural infections or immunizations. Clin Microbiol Infect. 2021.
- Assessment RR. Risk related to the spread of new SARS-CoV-2 variants of concern in the EU/EEA–first update. European Centre for Disease Prevention and Control An agency of the European Union. 2021.
- Challen R, Brooks-Pollock E, Read JM, Dyson L, Tsaneva-Atanasova K, Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern: matched cohort study. BMJ. 2021;372.
- World Health Organization. European Centre for Disease Prevention and Control/WHO Regional Office for Europe. Tuberculosis Surveillance and Monitoring in Europe. Geneva. 2013.
- Vogels CB, Breban M, Alpert T, Petrone ME, Watkins AE, Hodcroft E, et al. PCR assay to enhance global surveillance for SARS-CoV-2 variants of concern. MedRxiv.
- Korukluoglu G, Kolukirik M, Bayrakdar F, Ozgumus GG, Altas AB, Cosgun Y, et al. 40 minutes RT-qPCR Assay for Screening Spike N501Y and HV69-70del Mutations. BioRxiv. 2021.
- Brejová B, Hodorová V, Boršová K, Cabanová V, Reizigová L, Paul ED, et al. A SARS-CoV-2 variant with $\Delta $ H69/$\Delta $ V70 in the Spike protein circulating in the Czech Republic and Slovakia. ArXiv 2021.
Citation: Naaber P, Lahesaare A, Truu L, Soojarv A, Adamson A, Beljaev K, et al. (2021) Rapid Screening for Variants of Concern in Routine SARS-CoV-2 PCR Diagnostics. J Vaccines Vaccin. S13:005.
Copyright: © 2021 Naaber P, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

