Indexed In
- Academic Journals Database
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- China National Knowledge Infrastructure (CNKI)
- Scimago
- Ulrich's Periodicals Directory
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- MIAR
- University Grants Commission
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2020) Volume 11, Issue 4
Prevalence of Rotavirus Genotypes after Introduction of Monovalent Rotavirus Vaccine in Uzbekistan during 2014-2016
Nargiz Ibadullaeva1*, Musabaev1, Renat Latipov2, Said Sharapov1, Lyubov Lokteva1, Evgeniya Kazakova1, Elizaveta Joldasova1, Aziza Khikmatullaeva1, Malika Khodjaeva1, Umed Yusupov1 and Ilkhom Norbaev12WHO Country Office, Tashkent, Uzbekistan
Received: 01-Jul-2020 Published: 22-Jul-2020, DOI: 10.35248/2157-7560.20.11.419
Abstract
Introduction: Rotaviruses are one of the most leading causes of severe gastroenteritis in children less than five years
of age worldwide. This study describes prevalence of rotavirus A (RVA) genotypes in Uzbekistan during for the
period October-December in 2014 and2015-2016 after introduction of rotavirus vaccination.
Methods: In total, 17546 stool specimens testing for the presence of rotavirus antigen by EIA was performed by using
the Prospect Rotavirus Kit (Oxoid Ltd.UK). In total 318 EIA positive samples were randomly selected and genotyped
by using one-step conventional reverse transcription polymerase chain reaction (RT-PCR). RT-PCR was performed
using a Qiagen One-Step RT-PCR kit (Qiagen, Inc., Valencia, CA) and Rotavirus Genotyping Oligonucleotide
Primers (CDC, Atlanta).
Results: The results showed a change in the circulating genotypes towards the prevalence of the genotype G2P[4] and
a decrease in the prevalence of the genotype G1P[8].
Conclusion: The prevalence of the genotype G2P[4] is not necessarily due to vaccine escape, but can also occur in the
course of the natural fluctuation of RVA genotypes, both geographically and temporally and this tendency requires
further monitoring.
Keywords
Rotaviruses; Antigen; Vaccine; Enzyme immunoassay; RVA genotype
Introduction
Acute diarrheal diseases of childhood remain one of the important public health problems due to the high incidence rate, the development of severe forms, complications and high mortality. To date, the etiological structure of acute diarrheal diseases has changed significantly. Despite the fact that the etiological structure of acute diarrhea differs in different countries of the world, but still the dominant are diarrhea caused by viruses [1,2]. Rotaviruses are the most common cause of severe diarrhea in children less than 5 years of age around the world and globally 215 000 deaths associated with this infection have occurred in 2013 [3]. The genome of rotavirus group A (RVA) consists of 11 double-stranded RNA (dsRNA) segments that contains six structural (VP1, VP2, VP3, VP4, VP6, VP7) and six nonstructural (NSP1-NSP6) proteins [4]. Viral classification rotaviruses are based on the serological characteristics and sequence diversity of the VP7 and VP4 proteins determining G and P genotypes, respectively [4,5]. To date, much information has been accumulated on the genetic diversity of rotaviruses. At the present time 36 G genotypes and 51 P genotypes [6] have been identified, and the most widespread combinations of genotypes of RVA are G1P[8], G2P[4], G3P[8], G4P[8], G9P[8] and G12P[8] [7-9]. There are geographic differences in the prevalence of different rotavirus genotypes and the frequency of occurrence of these genotypes, which may change with time. Change in the landscape of genotypes can be observed after the introduction of vaccination. In the Republic of Uzbekistan since June 2014, vaccination against rotavirus disease (Rotarix GlaxoSmithKline Inc., Rixensart, Belgium) has been included into the national immunization calendar. To assess the effectiveness of vaccination, it is necessary to know the genetic spectrum of rotaviruses common in a given area. The choice of the pathogen detection methods plays an important role. In some studies RTPCR assays targeting gene segments of the RVA (VP4, VP6, VP7) have shown a higher sensitivity in the rate of RVA detection in comparison with EIA [1,10-12].
The aim of this study was to investigate the landscape of circulating RVA genotypes in Uzbekistan during October- December in 2014 and 2015-2016 after introduction of rotavirus vaccination.
Materials and Methods
Epidemiological surveillance and stool specimen collection
In 2014, sentinel epidemiological surveillance of acute cases of diarrhea in two hospitals of the Republic of Uzbekistan was organized on the basis of the WHO guidelines [13] for assessing the effectiveness of vaccination and preventing severe cases of rotavirus infection (city infectious diseases hospital No.4 in Tashkent city and the regional infectious diseases hospital in the Bukhara city). Epidemiological surveillance included children aged from 0 to 59 months who had a liquid stool not less than 3 times in the last 24 hours but not longer than 7 days. In sentinel surveillance included stool samples from every third child incoming to hospital with diarrhea. The stool samples were collected and held in a special container during the first 24 hours from the moment the patient enters the hospital and were stored at 2°C -8°C not more than 2 weeks before delivery to the laboratory. All samples were sent to the Reference Laboratory of the Research Institute of Virology and stored at minus 20° .On October-December 2014 and 2015-2016, 17546 stool samples were collected from children 5 years old hospitalized with acute gastroenteritis.
Enzyme immunoassay (EIA)
Stool samples testing for the presence of rotavirus antigen by EIA was performed by using the Prospect Rotavirus Kit (Oxoid Ltd.UK) according to the manufacturer's instructions.
Viral RNA extraction and RVA genotyping
Viral RNA was extracted from 10% stool suspensions prepared in phosphate buffered saline (PBS) using QIAamp Viral RNA Mini kit (Qiagen, GmbH, Hilden, Germany) according to the manufacturer's instructions. One-step conventional reverse transcription polymerase chain reaction (RT-PCR) was performed using a Qiagen One-Step RT-PCR kit (Qiagen, Inc., Valencia, CA) and Rotavirus Genotyping Oligonucleotide Primers (CDC, Atlanta). For the VP7 genotyping (G1, G2, G3, G4, G9, G12) the forward primer 9con1-L and the reverse primers 9T-1, 9T-2, 9T-3P, 9T-4, 9T-9B, G12S were used and for VP4 genotyping (P[4], P[6], P[8], P[9], P[10]) the forward primer con3 and the reverse primers 1T-1D, 2T-1, 3T-1, 4T-1, 5T-1 as described previously [14,15]. After denaturing RNA (5 min at 95°C), the RT-PCR reaction was carried out in the Applied Biosystems Thermal Cycler “Veriti”. The reaction conditions were: 30 min at 50°C for the RT step; 15 minutes at 95°C; 30 PCR cycles (30 seconds at 94°C, 30 seconds at 50°C and 45 seconds at 72°C); 7 minutes at 72°C; 4°C storage. Detection of amplification products was performed by gel electrophoresis in 3% of SeaKem ME Agarose (Lonza Inc, Rockland, ME) which was stained with GelRed (BiotiumInc) using Track it 100 bp DNA Ladder (Thermo Fisher Scientific) in 1X TAE buffer at 110 Volts for 90 min. VP7 (G) and VP4 (P) genotyping was carried out according the size of amplicons as described previously.
Results
In total, 17546 stool specimens were collected after introduction rotavirus vaccination on (October-December) 2014 and 2015-2016 from children<5 years of age, who were hospitalized with acute gastroenteritis in two hospitals in Uzbekistan (city infectious diseases hospital No.4 in Tashkent city and the regional infectious diseases hospital in Bukhara city). Antigen of RVA was detected by EIA in 2547/17546 (14.5%) samples. The results of EIA during the study period were as follows: (October- December) 2014 – 680/2466 (27.6%), 2015–899/7806 (11.5%), 2016 – 968/7274 (13.3%). Analysis of the results of our study by EIA showed a decrease in the rate of rotavirus infection [16]. In total 318 EIA positive samples were randomly selected and genotyped: 61 samples from (October-December) 2014, 185 samples from 2015 and 72 samples from 2016. The analysis of the obtained results shows that 313 (98.4%) samples were typeable for both, VP7 (G) and VP4 (P). G types were determined for 317 (99.7%), and 1 (0,3%) sample was untypable. P types were determined for 314(98.7%), and 4 (1.3%) were untypeable. Mixed genotypes i.e. more than one genotype founded in an individual sample were determined for 4 (1.3%).
In 2014-2016, genotype G2P[4] was the most common genotype and detected in 141 (44.3%) samples. Other the most common genotypes in 2014-2016 were identified in the following order: G9P[8] – 69 (21.7%), G4P[8] – 29 (9.1%), G1P[8] – 27 (8.4%), whereas, common genotypes G3P[8] was only detected in 2014. In 2014-2016, uncommon genotype G2P[6] was detected in 5 (1.6%) samples, genotype G9P[4] accounted for 34 (10.5%) samples in 2014-2015, and prevalence other genotypes are presented in the Table 1.
| Genotypes | Post vaccination period | |||||
|---|---|---|---|---|---|---|
| 2014 | 2015 | 2016 | ||||
| (n=61) | (n=185) | (n=72) | ||||
| n | % | n | % | n | % | |
| G1P[8] | 2 | 3.3 | 20 | 10.8 | 5 | 6.9 |
| G2P[4] | 26 | 42.6 | 72 | 38.9 | 43 | 59.7 |
| G2P[6] | 1 | 1.6 | 1 | 0.5 | 3 | 4.2 |
| G2P[8] | 0 | 0 | 2 | 1.1 | 0 | 0 |
| G4P[8] | 12 | 19.7 | 4 | 2.2 | 13 | 18.1 |
| G9P[4] | 10 | 16.4 | 24 | 12.9 | 0 | 0 |
| G9P[8] | 6 | 9.8 | 61 | 32.9 | 2 | 2.8 |
| Other1 | 1 | 1.6 | 0 | 0 | 1 | 1.4 |
| Mixed2 | 2 | 3.3 | 0 | 0 | 2 | 2.8 |
| Untypable3 | 1 | 1.6 | 1 | 0.5 | 3 | 4.2 |
1Other: genotypes which identified in isolated cases; 2Mixed: more than one genotype found in one individual sample; 3Untypable: one of genotypes (G or P) untypable.
Table 1: Prevalence RVA genotypes after the introduction of rotavirus vaccination
Discussion
According to the WHO, by the end of 2016, rotavirus vaccines have been introduced in 90 countries, and global coverage is estimated at 25%, and the efficacy and safety of these vaccines have been proven in many clinical trials [17-22].
In 2005-2009, in order to assess the burden of rotavirus infection in three countries of Central Asia (Uzbekistan, Kazakhstan, Kyrgyzstan) was conducted a prospective hospitalbased surveillance for rotavirus diarrhea. The results of these studies presented an epidemiological picture of rotavirus infection in Central Asia and showed a substantial rotavirus burden which can be prevented by vaccination [23]. The results of this large prospective study also formed the basis for the decision to introduce vaccination against rotavirus infection in the Republic of Uzbekistan since June 2014, and since January 2014 was organized sentinel surveillance of rotavirus infection. In Uzbekistan, rotavirus vaccination coverage reached 95%-99% [24].
Conducting molecular genetic studies allows us to determine the circulating genotypes characteristic of a particular region. The existing geographical differences in the prevalence of different rotavirus genotypes and the frequency of occurrence of these genotypes can vary from year to year. Results of studies of circulating genotypes in Uzbekistan conducted prior to the introduction of rotavirus vaccination in the period 2005 show the prevalence of the genotype G1P[8] – 52.0%, as well as the prevalence of genotypes G2P[4]-19.0% [16]. In neighboring Central Asian Republics i.e. in Kyrgyzstan and Kazakhstan in 2007-2009, the predominant genotypes were G1P[8], G2[4], and G3P[8]. In Kyrgyzstan, the dominant genotype was G1P[8]-51.0-64.7% of cases, and in Kazakhstan G2P[4] prevailed [25]. These research results show similarity in the prevalence of circulated genotypes in the period of time before vaccination in neighboring Republics. Similarly, predominated genotype G1P[8] in 2007-2012 were reported in African Region (AFR), Western Pacific Region (WPR), South East Asia Region (SEAR), Eastern Mediterranean Region (EMR), European Region (EUR), the only exception was American Region (AMR) where genotype G2P[4] was relatively more commonly prevalence [8].
Our results obtained after introduction of vaccination show a change in circulating RVA genotypes towards the prevalence of the genotype G2P[4] and a decrease in the prevalence of G1P[8] (Figure 1).
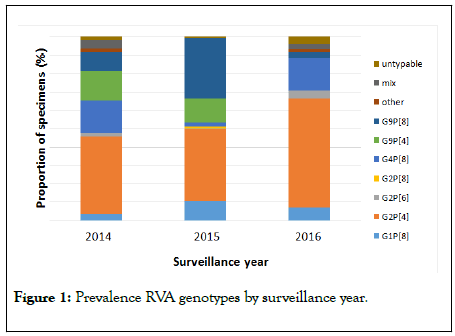
Figure 1: Prevalence RVA genotypes by surveillance year.
In 2014, G2P[4] was the predominant genotype-42.6%, and the occurrence of the genotype G1P[8] was 3.3%. But, in 2015, predominated two genotypes G2P[4]-38.9% and G9P[8]-32.9%, and the prevalence of G1P[8] was 10.8%. The predominant two genotypes G2P[4] and G9P[8] were also observed in post vaccination period in Scotland where a monovalent vaccine was used [26]. Whereas in Yemen where the monovalent vaccine was also used, after introduction vaccination the predominant genotypes were G1P[8] and G9P[8] [27]. Transient increase in the frequency of genotypes G9P[8] and G12P[8] were reported in some high vaccine coverage areas within the USA [8]. In 2016, G2P[4] was also the predominant genotype – 59.7%, and genotype G1P[8] was found in 6.9% of cases. An increase in the prevalence of G2P[4] was observed every year, whereas the circulation of the genotypes G4P[8], G9P[4], G9P[8] varied. After the introduction of monovalent rotavirus vaccine, such changes in the proportion of genotypes, i.e. the decrease in the prevalence of the genotype G1P[8] and the prevalence toward the genotype G2P[4] was also observed in Saudi Arabia [28]. Genotype G2P[4] was predominant in Belgium, Austria, Colombia after introduction rotavirus vaccination [29-31]. An increase of G2P[4] RVA isolates was also observed in Brazil after introduction of Universal Mass Vaccination (UMV) using the Rotarix vaccine [32]. However, the vaccine was efficacious in reducing the absolute number of natural RV infections/disease [33]. In Brazil before vaccine introduction and in neighboring countries of S America where UMV with RV vaccine has not been introduced, increases of G2P[4] RVA isolates were observed [34].
Thus, the relatively higher frequency of G2P[4] RVA isolates is not necessarily due to vaccine escape, but can also occur in the course of the natural fluctuation of RVA genotypes, both geographically and temporally. The prevalence of G2P[4] strains reported some countries after introduction Rotarix vaccine is unusual, and this issue requires further monitoring [8]. In order to find out whether there are differences between the strains of genotype G1P[8] that circulate in the post-vaccination period, further in-depth molecular genetic studies are needed.
Conclusion
Thus, analysis of the genotypes of rotaviruses will allow identifying regional genetic characteristics of the pathogen, monitoring the vaccination strategy as well as forecasting the epidemic situation and carrying out anti-epidemic measures.
Acknowledgments
This study was supported by Centers for Disease Control and Prevention (USA, Atlanta) and World Health Organization.
REFERENCES
- Latipov RR, Asilova MU, Musabaev EI, Rakhmanova JA. Pathogenic microflora in stool samples and clinical manifestation of acute diarrhea among young children in Uzbekistan. CA Cancer J Clin.2013; 1: 78-86.
- Liu L, Johnson HL, Cousens S, Perin J, Scott S, Lawn JE, et al. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. The Lancet. 2012; 379: 2151-2161.
- Tate JE, Burton AH, Boschi-Pinto C, Parashar UD. Global, regional, and national estimates of rotavirus mortality in children <5 years of age, 2000-2013: Clin Infect Dis. 2016; 62: 96-105.
- Abdulazeez AA. The histopathological effect of rotavirus in small intestine of mice isolated from wasit province. IJONS.2018;9: 16862- 16867.
- Iturriza-Gómara M, Kang G, Gray J. Rotavirus genotyping: keeping up with an evolving population of human rotaviruses. J ClinVirol. 2004; 31(4): 259-265.
- RCWG. Rotavirus Classification Working Group: RCWG. 2016. Available online: https://rega.kuleuven.be/cev/ viralmetagenomics/virus-classification/rcwg
- Patel MM, Steele D, Gentsch JR, Wecker J, Glass RI, Parashar UD. Real-world impact of rotavirus vaccination. Pediatr Infect Dis J.. 2011; 30: S1-5.
- Dóró R, László B, Martella V, Leshem E, Gentsch J, Parashar U, et al. Review of global rotavirus strain prevalence data from six years post vaccine licensure surveillance: is there evidence of strain selection from vaccine pressure? Infect Genet Evol. 2014;28: 446-461.
- Gómez MM, Resque HR, de Mello Volotão E, Rose TL, da Silva MF, Heylen E, et al. Distinct evolutionary origins of G12P [8] and G12P [9] group A rotavirus strains circulating in Brazil. Infect Genet Evol. 2014; 28: 385-388.
- Mijatovic-Rustempasic S, Tam KI, Kerin TK, Lewis JM, Gautam R, Quaye O, et al. Sensitive and specific quantitative detection of rotavirus A by one-step real-time reverse transcription-PCR assay without antecedent double-stranded-RNA denaturation. J Clin Microbiol. 2013; 51(9): 3047-3054.
- Pang XL, Joensuu J, Hoshino Y, Kapikian AZ, Vesikari T. Rotaviruses detected by reverse transcription polymerase chain reaction in acute gastroenteritis during a trial of rhesus-human reassortant rotavirus tetravalent vaccine: implications for vaccine efficacy analysis. J Clin Virol. 1999; 13: 9-16.
- Wilde J, Van R, Pickering L, Eiden J, Yolken R. Detection of rotaviruses in the day care environment by reverse transcriptase polymerase chain reaction. J Infect Dis. 1992;166: 507-511.
- World Health Organization. Generic protocol for monitoring impact of rotavirus vaccination on gastroenteritis disease burden and viral strains. World Health Organization; 2008.
- Das BK, Gentsch JR, Cicirello HG, Woods PA, Gupta A, Ramachandran M, et al. Characterization of rotavirus strains from newborns in New Delhi, India. J Clin Microbiol.1994; 32: 1820-1822.
- Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, et al. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992; 30: 1365-1373.
- Flem ET, Musabaev E, Juraev R, Kerin T, Gentsch J, Roger I, et al. Bresee. Rotavirus Gastroenteritis in Uzbekistan: Implications for Vaccine Policy in Central Asia. J Infect Dis. 2009; 200: 154–159.
- Bonkoungou IJ, Aliabadi N, Leshem E, Kam M, Nezien D, Drabo MK, et al. Impact and effectiveness of pentavalent rotavirus vaccine in children< 5 years of age in Burkina Faso. Vaccine. 2018; 36: 7170-7178.
- Ruiz-Palacios GM, Pérez-Schael I, Velázquez FR, Abate H, Breuer T, Clemens SC, et al. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N Engl JMed. 2006; 354: 11-22.
- Vesikari T, Matson DO, Dennehy P, Van Damme P, Santosham M, Rodriguez Z, et al. Safety and efficacy of a pentavalent human– bovine (WC3) reassortant rotavirus vaccine. N Engl JMed. 2006; 354: 23-33.
- Yoshikawa T, Matsuki T, Sato K, Mizuno M, Shibata M, Hasegawa S, et al. Impact of rotavirus vaccination on the burden of acute gastroenteritis in Nagoya city, Japan. Vaccine. 2018; 36: 527-534.
- Yen C, Tate JE, Hyde TB, Cortese MM, Lopman BA, Jiang B, et al. Rotavirus vaccines: current status and future considerations. Hum VaccinImmunother. 2014; 10: 1436-1448.
- Bar-Zeev N, Kapanda L, Tate JE, Jere KC, Iturriza-Gomara M, Nakagomi O, et al. Heyderman RS, French N. Effectiveness of a monovalent rotavirus vaccine in infants in Malawi after programmatic roll-out: an observational and case-control study. Lancet Infect Dis. 2015;15: 422-428.
- Latipov R, Utegenova E, Kuatbayeva A, Kasymbekova K, Abdykarimov S, Juraev R, et al. Epidemiology and burden of rotavirus disease in Central Asia. Int J Infect Dis. 2011;15: 464-469.
- World Health Organization. WHO vaccine-preventable diseases: monitoring system. Geneva: WHO. 2003.
- Vainio K, Latipov R, Utegenova E, Kasymbekova K, Juraev R, Asilova M, et al. Rotavirus genotype distribution in K yrgyzstan and K azakhstan. 2007–2009. Apmis. 2013;121: 447-455.
- Mukhopadhya I, Murdoch H, Berry S, Hunt A, Iturriza-Gomara M, Smith-Palmer A, et al. Changing molecular epidemiology of rotavirus infection after introduction of monovalent rotavirus vaccination in Scotland. Vaccine. 2017; 35: 156-163.
- Amood AL-Kamarany M, Al Areqi L, Mujally A, Alkarshy F, Nasser A, Jumaan AO. Diarrheal diseases hospitalization in Yemen before and after Rotavirus Vaccination. Scientifica. 2016.
- Al Ayed MS, Asaad AM, Qureshi MA, Hawan AA. Epidemiology of group A rotavirus infection after the introduction of monovalent vaccine in the National Immunization Program of Saudi Arabia. J Med Virol.2017; 89: 429-434.
- Braeckman T, Van Herck K, Meyer N, Pirçon JY, Soriano-Gabarró M, Heylen E, et al. Effectiveness of rotavirus vaccination in prevention of hospital admissions for rotavirus gastroenteritis among young children in Belgium: case-control study. BMJ. 2012; 345.
- Gilbert SC. Clinical development of Modified Vaccinia virus Ankara vaccines. Vaccine. 2013; 31: 4241-4246.
- PeláezCarvajal D, CotesCantillo K, PaterninaCaicedo A, Gentsch J, de la HozRestrepo F, Patel M. Characterization of rotavirus genotypes before and after the introduction of a monovalent rotavirus vaccine in Colombia. J Med Virol.2014; 86: 1083-1086.
- Gurgel RG, Bohland AK, Vieira SC, Oliveira DM, Fontes PB, Barros VF, et al. Incidence of rotavirus and all-cause diarrhea in northeast Brazil following the introduction of a national vaccination program. Gastroenterology. 2009;137: 1970-1975.
- Gurgel RQ, Alvarez AD, Rodrigues A, Ribeiro RR, Dolabella SS, Da Mota NL, et al. Incidence of rotavirus and circulating genotypes in Northeast Brazil during 7 years of national rotavirus vaccination. PLoS One. 2014; 9: 110217.
- Gómez MM, Carvalho-Costa FA, de Mello Volotão E, Rose TL, da Silva MF, Fialho AM, et al. Prevalence and genomic characterization of G2P [4] group A rotavirus strains during monovalent vaccine introduction in Brazil. Infect Genet Evol. 2014; 28 : 486-494.
Citation: Ibadullaeva N, Musabaev, Latipov R, Sharapov S, Lokteva L, Kazakova E, et al. (2020) Prevalence of Rotavirus Genotypes after Introduction of Monovalent Rotavirus Vaccine in Uzbekistan during 2014-2016. J Vaccines Vaccin.11:419. DOI: 10.35248/2157-7560.20.11.419
Copyright: © 2020 Ibadullaeva N, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

