Indexed In
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- Ulrich's Periodicals Directory
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Proquest Summons
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2023) Volume 0, Issue 0
Phosphorylation State of S6K1 is Redundant for its Blood Interaction with F Actin
Shafat A Latoo1* and Khurshid I Andrabi22Department of Biotechnology and Bioinformatics, University of Kashmir, Jammu and Kashmir, India
Received: 21-Aug-2023, Manuscript No. JBDT-23-22728; Editor assigned: 23-Aug-2023, Pre QC No. JBDT-23-22728 (PQ); Reviewed: 13-Sep-2023, QC No. JBDT-23-22728; Revised: 20-Sep-2023, Manuscript No. JBDT-23-22728 (R); Published: 27-Sep-2023, DOI: 10.4172/2155-9864.23.S3.011
Abstract
The evolutionarily conserved kinase (S6K1) that is actuated in response to certain stimulants like insulin, amino acids and several other growth factors has emerged as a major effector of growth and proliferation of blood cells. In certain types of cancers, the S6K1 a serine/threonine kinase is constantly actuated and acts as a downstream effector of the Akt/phosphatidylinositol 3-kinase pathway. S6K1 serves to cross-link actin filament and activates the Rho family of Guanosine Triphosphate (GTPases). We here present the evidence for domain-specific interaction of S6 kinase 1 (S6K1) with filamentous actin or F actin. We showed for the first time that the [ΔNH2-146/ΔCT240 a. acid] region of S6K1 is responsible for its discrete binding to F actin. We also demonstrate that S6K1 binding to filamentous actin is phosphorylation independent and not facilitated by any other protein rather than direct interaction and we couldn’t observe any interaction of S6K1 for monomeric actin (G actin). By a time course experiment, we could found that the kinetics of spontaneous actin polymerization is not affected by the presence of S6K1 rather it enforces stability in F actin by cross-linking it and rendering it more stable in the form of multifilament bundled actin. Using electron microscopy we found that these closely packed bundles were often somewhat curved, which could suggest pliable cross-linking. We further observe that S6 kinase 1 persists to show reactivity towards filamentous actin that stands unaltered amid deletions compromised for [ΔNH2-146/ΔCT104] or [ΔNH2-46]/ΔCT104] or [ΔNH2-146] or [ΔNH2-46] or [ΔCT104]. By computational study, we found that the [ΔNH2-146/ΔCT240 a. acid] region of S6K1 is rich in hydrophobic amino acids and holds primarily coiled-coil and alpha-helical structure which serves as a structural basis for some of the actin-binding proteins. These data together with the capability of S6K1 to attach filamentous actin indicate that binding is phosphorylation independent, direct, and facilitated by the [ΔNH2-146/ΔCT240 a. acid] region of S6K1.
Keywords
S6K1; F-actin; G-actin; Cancer; Cross-linking; Phosphorylation; Blood; Actin cytoskeleton
Abbreviations
S6K1: 40S Ribosomal Protein S6 Kinase 1; p70S6K: p70 Ribosomal S6 Kinase; HGF: Hepatocyte Growth Factor; mTOR: mammalian Target of Rapamycin; PI3K: Phosphoinositide-3-Kinase; F actin: Filamentous actin; G actin, Globular actin
Introduction
In human ovarian cancer, the ribosomal kinase p70 S6 (p70S6K) is routinely activated and functions as an effector of the Akt/ phosphatidylinositol 3-kinase signaling pathway [1-3]. Hepatocytes and epidermal growth factors including cytokines act as potent inducers of S6K1 in ovarian carcinoma or malignant epithelial ovarian tumors [4,5].
Continuous activation of S6K1 occurs much more frequently in malignant ovarian tumors than in benign lesions [6]. S6K1 has a well-known role in regulating blood cell survival and proliferation and may also play a role in other features of ovarian cancer progression, such as metastasis [5,7,8].
It has been observed that the rearrangement of the actin cytoskeleton is necessary and very important for cell migration [9]. A study by Chou et al. confirmed that p70S6K and Rho GTPases coexist in a comparable pathway [10]. P70S6K is activated by the fusion of Cdc42 and Rac1, which can be blocked by PI3K inhibitor (wortmannin) and mTOR inhibitor (mammalian Target of Rapamycin) [10].
Under the influence of growth factor stimulation, Berven et al. confirmed the role of p70S6K in the reorganization of the actin cytoskeleton through its colocalization with the actin arch and the Swiss3T3 fibroblast stress fibers at its anterior border [11]. Treatment with rapamycin, which inhibits p70S6K, may prevent the organization and elongation of actin stress fibers, suggesting that colocalization of p70S6K and stress fibers regulates this event [12].
In directed blood cell migration, as in the case of metastasis, p70S6K acts as a key and central regulator [13]. In the current study, we could demonstrate at the earliest domain-specific interaction of S6K1 with F actin and further show that binding of S6K1 with F actin is phosphorylation independent, direct, and facilitated by the [ΔNH2-146/ΔCT240 a. acid] region of S6K1.
Materials and Methods
Antibodies
Actin, antibiotics, Dulbecco's Modified Eagle Medium (DMEM), Fetal Bovine Serum (FBS), TNM-FH media (Sigma-Aldrich St Louis, MO), Rapamycin (Calbiochem, USA) and every single reagent was purchased from Sigma if not designated, anti (Glutathione-S- Transferase) GST (raised in the lab), the Polyvinylidene Fluoride (PVDF) transfer membrane ((Millipore Billerica, MA, USA), anti S6K (Santa Cruz Biotechnology, Inc.-Texas, USA), anti β actin (Epitomics, Inc.-Burlingame, California, USA), Highly cross- adsorbed Goat anti rabbit secondary antibody conjugated to IRDye 800CW (Li-Cor Biosciences, Lincoln, Nebraska, USA).
Plasmid constructs
Influenza virus N terminal Hemagglutinin (HA) epitope tagged Rattus S6 Kinase α1 (p85) (S6K1) and S6K (Δ2-46) (ΔCT140) cloned at Escherichia coli restriction (EcoR1) site in the pMT2 vector was a kind gift from Joseph Avruch (Harvard Medical School). Using the appropriate primers and the conventional Polymerase Chain Reaction (PCR) procedure, the pMT2 vector containing S6K and its truncated versions were amplified. The pGEX4T-2 vector's GST (Glutathione-S-transferase) coding sequence was in frame with the primers, which carried 5'-Bam H1 and 3'-Eco R1 in the amplified fragment.
S6K1 (p85): (5’- GGCGGATCCAGGCGAC GACGGAGGCGG GACGGCTTTTAC-3’ 5’-CGGGAATCCTCATAGATTCATAC GCAGGTGCTCTGGCCGTTT-3’)
S6K1 (Δ2-146): (5’- GGCGGATCCCTGGAGGAAGT AAAGCAT CCCTTCATTGTG-3’ 5’-CGGGAATCCTCATAGATTCAT ACGC AG GTGCTCTGGCCGTTT-3’)
S6K1 (ΔCT104): (5’- GGCGGATCCAG GCGACGACGGAGGC GGGACGGCTTTTAC-3’ 5’- CGGGAATTCTCATTTCAA GTACAGA TGGAGCCACATAT GTAA-3’)
S6K1 (Δ2-146/ΔCT104): (5’- GGCGGATCCCTGGAGGAAG TAA AGCATCCCTTCATTGTG-3’ 5’-CGGGAATTCT CATTTCAAGTACAGATGGAGCCACATATGTAA-3’)
S6K1 (Δ2-146/ΔCT240): (5’- GGCGGATCCCTGGAGGAAGTAA AGCATCCCTTCATTGTG-3’ 5’-CGGGAATTCTCATACAT TAATGCTCCCAGACT CCACCAATCC-3’)
S6K1 (Δ2-285/ΔCT104): (5’- GGCGGATCCT G ACATGCTGACTGGAGCACCTCCATTCAC-3’ 5 ’ - C G G G A A TTCTCATTTCAAGTACAGATGGAG CCACATATGTAA-3’)
S6K T412 E (Threonine replaced with Glutamic acid) using primer set by quick change Tm site directed mutagenesis kit (Santa Clara, CA) in accordance with the protocol stated in Zhang et al.
Template used: pMT2-S6K
Primers used:
• 5 ’ C T GGGTTTTGAATATGTGGCTCCATCTGTA CTTGAAAGTG3’
• 5’---GAGCCACATATTCAAAACCCAGAAAGACCTG GTTGGCACTTT---3’
S6K412E: S6K variant with a point mutation at amino acid 412 in the linker domain, where threonine is altered to glutamic acid to mimic phosphorylated residue.
Generation of recombinant viruses and cell culture
Recombinant viruses were generated by ligating EcoR1 digested fragment from pMT2-S6KWT and various mutants in pVL-1392 and sequenced to check orientation and then co-transfected with linearized Baculo-viral Deoxyribo Nucleic Acid (DNA) using Baculo-gold kit (BD Biosciences, San Diego, California, USA) in accordance with the manufacturer’s instructions. Plaque assay was performed strictly in line with manufacturer’s instructions to calculate viral titre. Insect cells (SF9) were grown in serum free TNM-FH medium, seeded at 2 × 106cells per 60 mm tissue plate, and transfected with individual viruses for each of the aforementioned construct at a Multiplicity of Infection (MOI) of ≤ 1 for 58-60 hours.
Expression and purification of proteins
The BL21-DE3 pLysS host strain (Sigma) was transformed with the constructs pGEX4T2 (GST-S6) and the parental control plasmid pGEX4T2, and fusion protein expression was induced for 16 hours at 37°C with 1 mM Isopropyl β- d-1-thiogalactopyranoside (IPTG). Cell lysates were made using a standard blood cell lysis approach after bacteria were harvested at 5,500 rpm for 15 minutes at 4°C. Cells were lysed in a buffer containing Tris-Cl (20 mM), EDTA (2 mM), NaCl (150 mM), lysozyme (1 mg/mL), and phenylmethylsulfonyl fluoride (PMSF) (1 mM) at pH 6.8 and incubated on ice for 30 minutes. Next, sonication (5 sec pulses, 5 sec rest), and centrifugation for 15000 rpm for 30 minutes at 4°C were performed. Assays for the actin cross-linking of pre-cleared lysates were performed.
Actin cross-linking assay
Pre-cleared blood cell lysates of different S6K1 truncation versions were incubated with 4.5μM freshly produced F-actin for 1 hour at room temperature before being centrifuged at 14000 g for 1 minute to sediment bundled F-actin; the linear, unbundled actin remained in the supernatant. Separate pellet and supernatant fractions were collected, and the pellet fractions were washed three times in 1 × PBS. Sodium Dodecyl-Sulfate Polyacrylamide Gel Electrophoresis (SDS PAGE) was used to analyse equal amounts of pellet and supernatant, followed by Western blotting, or the gel was stained with Coomassie Blue.
G-actin (Globular Protein) with a molecular weight of 43 Kilo Dalton (KD) was obtained commercially from Sigma Aldrich. G actin stocks were prepared in sterile water, filter sterilized using 0.25 micron bacterial filters at a final concentration of 4.5μM, and kept at -20°C. Actin was prepared in a buffer containing 5 mM Tris pH 8.0, 0.2 mM Adenosine Triphosphate (ATP), 0.2 mM Calcium Chloride (Cacl2), 0.5 mM Dithiothreitol (DTT), or 0.5 mM β-mercaptoethanol for the binding assay and centrifuged at 20,000 g (14000 rpm) for 10 minutes at 4°C.The monomeric actin supernatant is ready for polymerization.
The addition of 50 mM Potassium chloride (KCL), 1 mM ATP, and 2 mM Magnesium chloride (Mgcl2) stimulates actin polymerization. Polymerization takes place for 1 hour at room temperature. Polymerized actin which is F-actin (Filamentous actin) is ready for actin cross-linking or cross-linking assay.
Western blotting
Proteins were loaded onto a 12% SDS-PAGE gel, run, then transferred to PVDF membranes by wet blotting at 70V continuous current for 90 minutes for Western blot analysis. Membranes were washed three times in 1 × PBS (Phosphate-Buffered Saline) with 0.05% tween 20 for ten minutes once and five minutes twice before being blocked overnight with ODYSSEY blocking buffer (LI-COR biotechnologies USA) or 3% nonfat milk in 1 × PBS. Primary antibodies were diluted 1:200 (anti-β actin; Epitomics), 1:250 (anti S6K; Santa Cruz Biotechnologies Inc, USA), and 1:10000 (GST antisera) in 0.5% blocking solution or 3% Bovine Serum Albumin (BSA) in 1 × PBS and incubated overnight at 4°C or for several hours at room temperature on a rocking plate. After three washes with PBS-T (PBS containing 0.05% Tween 20), the membranes were incubated for 1 hour at room temperature with a 1:10,000 dilution of an Infrared dye-conjugated secondary goat anti-rabbit antibody (800 CW). After washing with PBS-T, antibody binding was observed using the LICOR ODYSSEY infrared system. Blots were stripped in stripping buffer (10 mM glycine, 2% SDS, pH 2.0) and incubated at room temperature for 30 minutes with intermittent shaking for re-probing. Membranes were washed twice in Phosphate-Buffered Saline/Tween (PBS-T) at room temperature for 10 minutes each time with significant volumes of washing buffer. The membranes were then blocked and treated as previously described.
Chemical fixation of S6K1 [Δ2-146/ΔCT240]-F actin complexes
An aliquot of the pre-clear lysates of S6K1 [Δ2-146/ΔCT240] was mixed with an aliquot of 50 μl of F actin containing polymerization induced buffer (1 mM ATP, 2 mM Mgcl2 and 50 mM KCL). It was fixed with gultaraldehyde at a final concentration of 0.25% after 1 hour of incubation at room temperature.
Electron microscopy
5 μL aliquots of either fixed or unfixed S6K1 [Δ2-146/ΔCT240]-F actin complexes or F actin alone incubation combinations, primed as described above were adsorbed for 1 minute onto glowdischarged carbon-coated collodion films on copper grids. One or three drops of distilled water or buffer were used to wash some of the grids. Excess liquid was drained using filter paper, and the grids were negatively stained by progressively laying them on three drops of 2% uranyl acetate for 10 seconds each. Excess liquid was drained using filter paper, then suction with a capillary applied to the grid's edge, which was then left to air dry. Images were captured with a colony collapse disorder (CCD) camera attached to a HITACHI S-3000H (SEM) (HITACHI Science Systems, ltd Japan).
Computational study
The computational study of [ΔNH2-146/ΔCT240] S6K1 was performed by submitting its amino acid sequence into an on-line platform I-TASSER server.
Results
Domain specific interaction of S6K1 with F actin
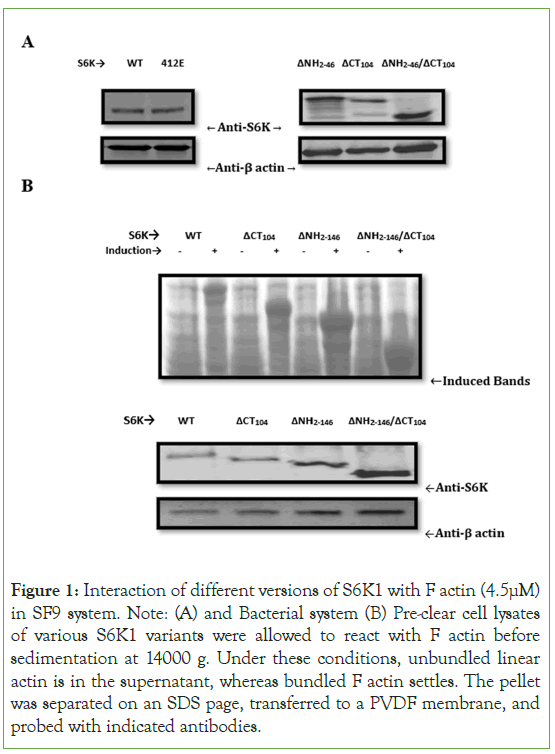
To better understand the mechanism of S6K1 involvement in actin cytoskeleton signaling, we conducted a co-sedimentation assay to see if S6K1 binds actin filament in vitro Figure 1. As illustrated in Figure1A, we expressed S6K1[WT, 412E] and its truncated versions in SF9 system and observe that S6K1 along with its truncated versions co-sediment with F actin with a binding potential more towards the catalytic domain truncated version of S6K1[ΔNH2-146/ΔCT104]. To validate our result we expressed S6K1 and its truncated versions in bacterial system which showed same result as in SF9 system (Figure 1B). We also tried to find out interaction of S6K1 and its different versions with G actin or monomeric actin but couldn’t observe any such interaction. The fact that no additional proteins have been associated suggests that S6K1 is directly bound to F actin in the bacterial system (Figure 1B).
Figure 1: Interaction of different versions of S6K1 with F actin (4.5μM) in SF9 system. Note: (A) and Bacterial system (B) Pre-clear cell lysates of various S6K1 variants were allowed to react with F actin before sedimentation at 14000 g. Under these conditions, unbundled linear actin is in the supernatant, whereas bundled F actin settles. The pellet was separated on an SDS page, transferred to a PVDF membrane, and probed with indicated antibodies.
Role of S6K1 in actin polymerization
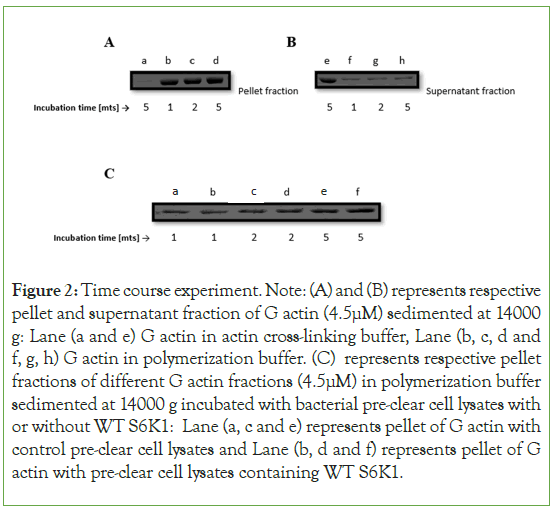
To investigate the kinetics of S6K1-induced actin bundle generation Figure 2, We did a time course experiment of WT S6K1 with actin and found that the presence of S6K1 doesn’t help in polymerization of G actin at all but it is induced by the polymerization buffer (Figure 2A,2B). It seems polymerization of actin is a very fast event reaching almost steady state within 5 minutes of its induction with polymerization buffer (Figure 2C) indicating that polymerization and bundling of actin is a synchronous event and thus providing an indication that S6K1quickly binds to actin filaments.
Minimal region of S6K1 responsible for binding to actin
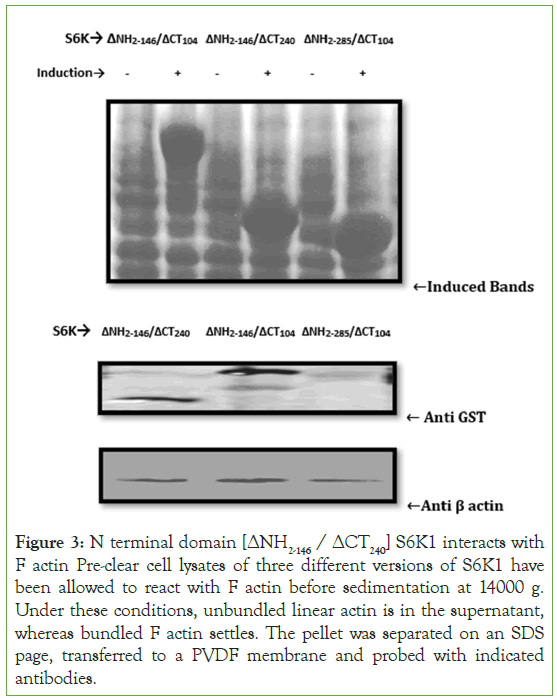
To identify which domain of S6K1 interacts with the actin, a chain of truncation variants has been generated and investigated by using cosedimentation assay. Due to the higher binding efficiency of [ΔNH2-146/ΔCT104] S6K1 and due to its predominant α helical structure has made us to focus on this domain of S6K1 so as to elucidate its functional significance in the binding kinetics of S6K1 and actin, we split [ΔNH2-146/ΔCT104] S6K1 into two halves corresponding to its N terminal halve comprising of [ΔNH2-146/ΔCT240] S6K1 and C terminal halve comprising of [ΔNH2-285/ΔCT104] S6K1 and expressed both of these constructs in bacterial system. We observed that N terminal halve [ΔNH2- 146/ΔCT240] of S6K1 shows binding specificity for its actin partner while as C terminal [ΔNH2-285/ΔCT104] lacks binding specificity (Figure 3). These findings indicate that S6K1's N terminal domain is the primary binding domain. It also narrowed down the minimal region in S6K1 which is responsible for binding to actin.
Figure 2: Time course experiment. Note: (A) and (B) represents respective pellet and supernatant fraction of G actin (4.5μM) sedimented at 14000 g: Lane (a and e) G actin in actin cross-linking buffer, Lane (b, c, d and f, g, h) G actin in polymerization buffer. (C) represents respective pellet fractions of different G actin fractions (4.5μM) in polymerization buffer sedimented at 14000 g incubated with bacterial pre-clear cell lysates with or without WT S6K1: Lane (a, c and e) represents pellet of G actin with control pre-clear cell lysates and Lane (b, d and f) represents pellet of G actin with pre-clear cell lysates containing WT S6K1.
Figure 3: N terminal domain [ΔNH2-146 / ΔCT240] S6K1 interacts with F actin Pre-clear cell lysates of three different versions of S6K1 have been allowed to react with F actin before sedimentation at 14000 g. Under these conditions, unbundled linear actin is in the supernatant, whereas bundled F actin settles. The pellet was separated on an SDS page, transferred to a PVDF membrane and probed with indicated antibodies.
Phosphorylation independent binding of S6K1 with F actin
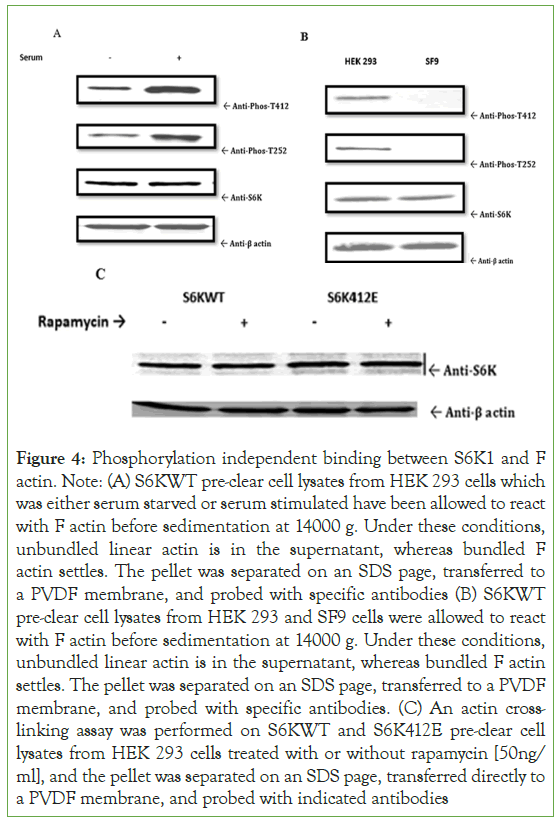
To investigate if S6K1 binding with F actin is phosphorylation dependent Figure 4, we couldn’t see any difference in binding under serum starved or serum stimulated conditions of S6KWT for F actin in HEK 293 system, which was again reconfirmed by looking at the binding kinetics of S6KWT for F actin in HEK 293 and SF9 system, and further confirmed by looking at the binding interaction of rapamycin treated or untreated one’s of S6WT and S6K412E in HEK 293 cells for F actin, which suggest that this binding phenomenon is not governed by phosphorylation as such, it happens in a manner independent of phosphorylation (Figures 4A-4C).
Figure 4: Phosphorylation independent binding between S6K1 and F actin. Note: (A) S6KWT pre-clear cell lysates from HEK 293 cells which was either serum starved or serum stimulated have been allowed to react with F actin before sedimentation at 14000 g. Under these conditions, unbundled linear actin is in the supernatant, whereas bundled F actin settles. The pellet was separated on an SDS page, transferred to a PVDF membrane, and probed with specific antibodies (B) S6KWT pre-clear cell lysates from HEK 293 and SF9 cells were allowed to react with F actin before sedimentation at 14000 g. Under these conditions, unbundled linear actin is in the supernatant, whereas bundled F actin settles. The pellet was separated on an SDS page, transferred to a PVDF membrane, and probed with specific antibodies. (C) An actin crosslinking assay was performed on S6KWT and S6K412E pre-clear cell lysates from HEK 293 cells treated with or without rapamycin [50ng/ ml], and the pellet was separated on an SDS page, transferred directly to a PVDF membrane, and probed with indicated antibodies
Role of minimal region of S6K1 on actin filament bundling
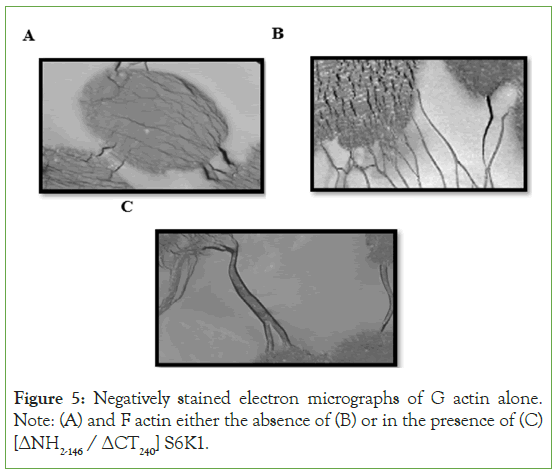
We used electron microscopy on negatively stained actin filaments to directly examine the effect of [ΔNH2-146 / ΔCT240] S6K1 on actin filament bundling. In the absence of [NH2-146/ΔCT240] S6K1, actin filaments formed a homogeneous network of fine filaments, as illustrated in (Figure 5B). However, higher order structures were found when actin was polymerized in the presence of [ΔNH2-146 / ΔCT240] S6K1 (Figure 5C). Despite the presence of single actin filaments, the majority of actin filaments were recruited into thick and lengthy actin bundles, indicating the cross-linking activity of [ΔNH2-146 / ΔCT240]. These tightly packed bundles were frequently slightly bent, indicating flexible cross-linking.
Figure 5: Negatively stained electron micrographs of G actin alone. Note: (A) and F actin either the absence of (B) or in the presence of (C) [ΔNH2-146 / ΔCT240] S6K1.
Computational study of minimal region of S6K1
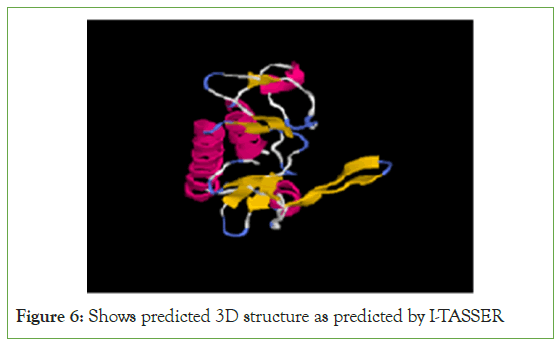
Bioinformatics study was carried out in order to further confirm [ΔNH2-146/ΔCT240 a. acid] S6K1 region; it was observed that this region contains predominantly hydrophobic amino acids and secondary structure prediction of this region has revealed predominant α helical and coiled coil regions which serves as the structural basis for some of the actin binding proteins (Figure 6).
Figure 6: Shows predicted 3D structure as predicted by I-TASSER
Discussions
In this study, we identified the S6 kinase 1 (S6K1) domain as a novel actin cytoskeleton regulator and demonstrated that it is essential for the cross-linking of F-actin (filamentous actin) and the induction of filamentous actin bundles. Numerous aspects of this function are important to remember. We demonstrated at an earliest the domain-specific interaction of S6K1 with filamentous actin (F-actin), with the binding potential tending towards the catalytic domain truncated version of S6K1 [ΔNH2-146 / ΔCT104]. This observation is consistent with the paradigm provided for S6K1 activation [14]. According to this model, the protein kinase exists in two states: inactive and active. When S6K1 is inactive, its carboxyl-terminal autoinhibitory domain, that shares sequence similarities with the S6 protein's substrate region, can behave as a pseudosubstrate and interact with its N-terminus. The removal of inhibition exerted by the autoinhibitory domain, according to the aforementioned idea, initiates S6K1 activation [15]. To fully activate the kinase, eight or more serine or threonine residues are phosphorylated at the autoinhibitory domain, the linker area, and subsequently the catalytic domain [14,16-20].
As a result, the higher activity of the different truncated versions of S6K1 relative to full-length S6K1 could simply be owing to the steric freedom acquired by the truncated versions of the enzyme, which is generally achieved by activating phosphorylations. In the case of the truncated version of the catalytic domain of S6K1 [ΔNH2- 146/ΔCT104], the binding site for the substrate is fully exposed, resulting in it having higher binding kinetics with its interaction partner F-actin, which is not the case for the full-length version, where this binding site for actin remains less exposed and hidden in the core of the protein and has slightly lower binding kinetics with F-actin, as determined by protein confirmation.
Actin cross-linking assays of preclear lysates of different S6K1 versions expressed in SF9 (insect cell line) and in a bacterial (prokaryotic) system have shown that the interaction is primarily with filamentous actin rather than monomeric actin (G-actin), which is well supported by a similar study by the group AST Wong et al [13]. The binding of S6K1 and F-actin is not facilitated by other proteins but is a direct interaction, as shown by the actin cross-linking experiment performed in a bacterial system. S6K1 was always found in the pellet, implying that pelleting is caused by F-actin binding. The absence of additional proteins suggests a direct relationship between S6K1 and actin.
The likelihood of a bodily interaction between S6K1 and actin indicates S6K1 may influence actin polymerization kinetics. In contrast to other activities shared by these proteins, S6K1 had no effect on the rate or extent of polymerization, implying that actin polymerization is not a general characteristic of S6K1. It appears that the polymerization of G-actin is a very rapid event, reaching a near steady state within 5 min of induction with polymerization buffer, suggesting that actin polymerization and bundling is a synchronous event, as observed by us and well supported by the light scattering experiment of the group AST Wong et al [13]. Actin filaments are constantly assembling and disassembling. For that reason, at steady state, each filament population contains a small but finite quantity of actin monomers, as demonstrated by the time course experiment [21,22]. Given the higher binding potential of [ΔNH2-146/ΔCT104] S6K1 and its predominant α-helical structure, we focused on this kind of domain to elucidate its functional significance for the binding kinetics of S6K1 and actin [23]. We demonstrated that [ΔNH2-146/ΔCT240] S6K1 is the most important binding domain. Furthermore, the minimal region in S6K1 responsible for binding to actin was narrowed down. Mutagenesis studies reveal that one of the neuronal proteins neurabin interacts with S6K1 via its PDZ domain in the middle of the protein [24].
This interaction is an example of heterotypic interaction of a PDZ region to the C-terminus of a substrate protein S6K1 (amino acids 332-502). S6K1 has no similarity with proteins containing a PDZ domain, and deletion of the C-terminal five amino acids of S6K1 abolishes binding. Neurabin was identified as an F-actin cross-linking protein by Nakanishi et al [25]. Neurabin interacts to F-actin through a distinct N-terminal domain (amino acids 1-144), which guides the protein inside the cytoskeletal region. This actinbinding region is separated from neurabin's PDZ domain, allowing actin and S6K1 to interact on their own, validating the sort of unique interaction we found between F-actin and (amino acids 147- 285) of S6K1 in the literature.
Bioinformatics study was carried out in order to further confirm this novel region; it was observed that this region contains predominantly hydrophobic amino acids and prediction of the secondary structure of this region revealed predominantly α-helical and coiled-coil regions, which serve as the structural basis for some known actin-binding proteins. Actin subdomains 1 and 3's hydrophobic cleft is thought to be a "hot spot" for Fand G-actin binding proteins. This cleft's conformation is such that it preferentially contacts the binding partner's α-helix, which is distinguished by the presence of several conserved and exposed side chains that are hydrophobic [26]. Aside from the α-helices of gelsolin, ciboulot, and albumin D-box Binding Protein (DBP), which bind to the aforementioned cleft in their corresponding structures with actin, the α-helix in actin's D-loop and α-helix 3 of Actin-depolymerization Factor (ADF)/cofilin have been proposed as well to bind to this cleft, and the Hematopoietic lineage cellspecific protein 1(HS1) coiled-coil domain has been seen to bind to F-actin [27-30].
The in vitro interaction between S6K1 with F-actin was not modulated by S6K1 phosphorylation, but occurred irrespective of phosphorylation, implying a phosphorylation-independent mechanism for S6K1 and F-actin binding.
We also used electron microscopy to see F-actin bundling. Actin filaments produced a homogeneous meshwork of fine filaments in the absence of [ΔNH2-146 / ΔCT240] S6K1, however there was no bundling. In its presence, however, robust multifilament bundles were plainly apparent. These tightly packed bundles were frequently somewhat bent, implying flexible cross-linking. The biological purpose of such an interaction is an intriguing element. The discovery that S6K1 both cross-links and stabilizes actin filaments suggests that S6K1 may have a role in modulating actin dynamics. Actin bundling is also regulated by several key actinbinding proteins, including fimbrin, α-actinin, and fascin [31,32]. Since S6K1 is known to play a function in protein synthesis, this connection may be critical for the production of local proteins required for the propagation of the migration reaction. β-actin messenger Ribonucleic acid (mRNA), for example, has been demonstrated to be localized within the leading lamella, where active translation is necessary for the migration of blood cells [33,34].
Conclusion
In conclusion, the current study identifies a new S6K1 domain and its role in regulating the dynamics of actin cytoskeleton. The S6K1 gene is typically active in ovarian cancer and other malignancies, and it plays an important role in various cancer-related processes, such as metastasis. The use of chemical inhibitors, siRNAs, and natural products to target this actin-binding region of S6K1 may provide a beneficial new route for cancer therapy and open up new areas of research in S6K1 biology.
These findings, combined with S6K1's capacity to bind to F actin, suggest that interaction is direct, phosphorylation independent, and aided by the [ΔNH2-146 / ΔCT240 a. acid] region of S6K1.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Acknowledgments
Dr. Joseph Avruch, Mass. General Hospital, Harvard Medical School, Boston-MA, generously donated HA-S6Kα1, [NH2-46/ CT140] S6Kα1. The authors also wish to acknowledge Dr Mushtaq Ahmad Beigh for the viral constructs and intellectual feedback. Aejaz-ul-Noor, Mehvish Showkat, Sheikh Tahir Majeed and Qussin Basharat for helpful discussion.
Declarations
Authors' contributions
SAL and KIA conception and layout, SAL data collecting, SAL and KIA data analysis and interpretation, SAL and KIA drafted the manuscript, and SAL and KIA revised it critically for essential intellectual quality. The version of the manuscript to be published has been approved by SAL and KIA.
Funding
Junior Research Fellowship in favor of SAL No: F (BTCH-Prj) KU/12/PB from the Department of Biotechnology (DBT), Infrastructural Grant under the FIST programme, and Special Assistance grant under the SAP programme from the Department of Science and Technology (DST) and the University Grants Commission (UGC) India.
Availability of data and materials
The corresponding author has access to all data related to this manuscript.
References
- Shayesteh L, Lu Y, Kuo WL, Baldocchi R, Godfrey T, Collins C. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet. 1999; 21 (1): 99–102.
[Crossref] [Google Scholar] [PubMed]
- Philp AJ, Campbell IG, Leet C, Vincan E, Rockman SP, Whitehead RH, et al. The phosphatidylinositol 3′-kinase p85α gene is an oncogene in human ovarian and colon tumors. Cancer Research. 2001;61(20):7426-7429.
[Crossref] [Google Scholar] [PubMed]
- Altomare DA, Wang HQ, Skele KL, Rienzo AD, Klein-Szanto AJ, Godwin AK. AKT and mTOR phosphorylation is frequently detected in ovarian cancer and can be targeted to disrupt ovarian tumor cell growth. Oncogene. 2004; 23(34): 5853–5857.
[Crossref] [Google Scholar] [PubMed]
- Liu LZ, Hu XW, Xia C, He J, Zhou Q, Shi X, et al. Reactive oxygen species regulate epidermal growth factor-induced vascular endothelial growth factor and hypoxia-inducible factor-1alpha expression through activation of AKT and P70S6K1 in human ovarian cancer cells. Free Radic Biol Med. 2006;41 (10): 1521–1533.
[Crossref] [Google Scholar] [PubMed]
- Zhou HY, Wong AS. Activation of p70S6Kinduces expression of matrix metalloproteinase 9 associated with hepatocyte growth factor-mediated invasion in human ovarian cancer cells. Endocrinology. 2006;147 (5): 2557–2566.
- Castellvi J, Garcia A, Rojo F, Ruiz‐Marcellan C, Gil A, Baselga J, et al. Phosphorylated 4E binding protein 1: A hallmark of cell signaling that correlates with survival in ovarian cancer.Cancer. 2006;107 (8): 1801–1811.
- Wong AS, Roskelley CD, Pelech S, Miller D, Leung PC, Auersperg N. Progressive changes in Met-dependent signaling in a human ovarian surface epithelial model of malignant transformation .Exp Cell Res. 2006;299 (1): 248–256.
[Crossref] [Google Scholar] [PubMed]
- Pon YL, Zhou HY, Cheung AN, Ngan HY, Wong AS. p70 S6 kinase promotes epithelial to mesenchymal transition through snail induction in ovarian cancer cells. Cancer Res. 2008 Aug 15;68(16):6524-6532.
[Crossref] [Google Scholar] [PubMed]
- Pantaloni D, Clainche CL, Carlier MF. Mechanism of actin-based motility. Science. 2001;292(5521):1502-1506.
[Crossref] [Google Scholar] [PubMed]
- Chou MM, Blenis J. The 70 kDa S6 kinase complexes with and is activated by the Rho family G proteins Cdc42 and Rac1. Cell. 1996;85(4):573-583.
- Crouch MF. Regulation of thrombin-induced stress fibre formation in Swiss 3T3 cells by the 70-kDa S6 kinase. Biochem Biophys Res Commun. 1997;233(1):193-199.
[Google Scholar] [PubMed]
- Berven LA, Willard FS, Crouch MF. Role of the p70S6K pathway in regulating the actin cytoskeleton and cell migration. Exp Cell Res. 2004;296(2):183-195.
[Crossref] [Google Scholar] [PubMed]
- Lp CKM, Cheung ANY, Ngan HYS, Wong AST. p70 S6 kinase in the control of actin cytoskeleton dynamics and directed migration of ovarian cancer cells. Oncogene. 2016;16(1):1-9.
[Crossref] [Google Scholar] [PubMed]
- Pullen N, Thomas G. The modular phosphorylation and activation of p70s6k. FEBS Lett. 1997;410(1):78-82.
[Crossref] [Google Scholar] [PubMed]
- Weng QP, Kozlowski M, Belham C, Zhang A, Comb MJ, Avruch J. Regulation of the p70 S6 kinase by phosphorylation in vivo. Analysis using site-specific anti-phosphopeptide antibodies. J Biol Chem. 1998; 273 (26): 16621-16629.
[Crossref] [Google Scholar] [PubMed]
- Alessi DR, Kozlowski MT, Weng QP, Morrice N, Avruch J. 3-Phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro. Curr Biol. 1998;8(2):69-81.
[Crossref] [Google Scholar] [PubMed]
- Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci U S A. 1998;95(4):1432-1437.
[Crossref] [Google Scholar] [PubMed]
- Dennis PB, Pullen N, Kozma SC, Thomas G. The principal rapamycin-sensitive p70(s6k) phosphorylation sites, T-229 and T-389, are differentially regulated by rapamycin-insensitive kinase kinases. Mol Cell Biol. 1996;16 (11): 6242-6251.
- Pearson RB, Dennis PB, Han JW, Williamson NA, Kozma SC, Wettenhall RE. The principal target of rapamycin-induced p70s6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. EMBO J. 1995; 14: 5279-1587.
- Han JW, Pearson RB, Dennis PB, Thomas G. Rapamycin, wortmannin, and the methylxanthine SQ20006 inactivate p70s6k by inducing dephosphorylation of the same subset of sites. J Biol Chem. 1995;270(36): 21396-21403.
- Kasai M, Asakura S, Oosawa F. The G-F equilibrium in actin solutions under various conditions. Biochim Biophys Acta. 1962;57(1):13-21.
[Crossref] [Google Scholar] [PubMed]
- Oosawa F, Asakura S, Hotta K, Imai N, Ooi T. G-F transformation of actin as a fibrous condensation. J Polymer Sci. 1959;26: 323-330.
[Crossref]
- Sunami T, Byrne N, Diehl RE, Funabashi K, Hall DL, Ikuta M, et al. Structural basis of human p70 ribosomal S6 kinase-1 regulation by activation loop phosphorylation. J Biol Chem. 2010; 285: 4587–4594.
- Burnett PE, Blackshaw S, Lai MM, Qureshi IA, Burnett AF, Sabatini DM, et al. Neurabin is a synaptic protein linking p70 S6 kinase and the neuronal cytoskeleton. Proc Natl Acad Sci U S A. 1998; 95 (14) 8351-8356.
[Crossref] [Google Scholar] [PubMed]
- Nakanishi H, Obaishi H, Satoh A, Wada M, Mandai K, Satoh K, et al. Neurabin: A novel neural tissue–specific actin filament–binding protein involved in neurite formation. J Cell Biol. 1997;139(4):951-961.
[Crossref] [Google Scholar] [PubMed]
- Dominguez R. Actin-binding proteins–a unifying hypothesis. Trends Biochem Sci. 2004;29(11):572-578.
[Crossref] [Google Scholar] [PubMed]
- McLaughlin PJ, Gooch JT, Mannherz HG, Weeds AG. Structure of gelsolin segment 1-actin complex and the mechanism of filament severing. Nature. 1993;364(6439):685-692.
- Otterbein LR, Cosio C, Graceffa P, Dominguez R. Crystal structures of the vitamin D-binding protein and its complex with actin: Structural basis of the actin-scavenger system. Proc Nat Aca Sci. 2002;99(12):8003-8008.
- Hertzog M, Van Heijenoort C, Didry D, Gaudier M, Coutant J, Gigant B, et al. The β-thymosin/WH2 domain: Structural basis for the switch from inhibition to promotion of actin assembly. Cell. 2004;117(5):611-623.
- Hao JJ, Zhu J, Zhou K, Smith N, Zhan X. The coiled-coil domain is required for HS1 to bind to F-actin and activate Arp2/3 complex. J Biol Chem. 2005;280(45):37988-37994.
[Crossref] [Google Scholar] [PubMed]
- Holmes GR, Goll DE, Suzuki A, Robson RM, Stromer MH. Effect of trypsin on rabbit skeletal muscle α-actinin. Biochim Biophys Acta. 1976;446(2):445-456.
[Crossref] [Google Scholar] [PubMed]
- Hanein D, Matsudaira P, DeRosier DJ. Evidence for a conformational change in actin induced by fimbrin (N375) binding. J Cell Biol.1997;139(2):387-396.
- Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J Cell Biol. 1994;127(2):441-451.
[Crossref] [Google Scholar] [PubMed]
- Latham Jr VM, Kislauskis EH, Singer RH, Ross AF. Beta-actin mRNA localization is regulated by signal transduction mechanisms. J Cell Biol. 1994;126(5):1211-1219.
[Crossref] [Google Scholar] [PubMed]
Citation: Latoo SA, Andrabi KI (2023) Phosphorylation State of S6K1 is Redundant for its Blood Interaction with F Actin. J Blood Disord Transfus. S3.011.
Copyright: © 2023 Latoo SA et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.