Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- CiteFactor
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research - (2022) Volume 13, Issue 3
Phenotypic and Phylogenetic Analyses of Residual Bacteria in Effective Microorganism-Fermented Cucurbitacin Phytonematicide
Phatu W. Mashela* and Kgabo M. PofuReceived: 13-Apr-2022, Manuscript No. JPPM-22-16109; Editor assigned: 15-Mar-2022, Pre QC No. JPPM-22-16109 (PQ); Reviewed: 29-Apr-2022, QC No. JPPM-22-16109; Revised: 06-May-2022, Manuscript No. JPPM-22-16109 (R); Published: 16-May-2022
Abstract
Information on residual bacteria that keep cucurbitacin phytonematicides aseptic for extended periods is not available. At 35 weeks after initiating fermentation, samples were collected from Nemafric-BL phytonematicide, with the cultured strain subjected to various phenotypic and phylogenetic tests. Phenotypic tests characterized the test residual bacteria as Gram-positive Lactobacillus strain, with energy-efficient homolactic fermentative process, but without CO2 as an end-product. Phylogenetic analysis using ZymoBIOMICSTM DNA Miniprep Kit for Gram- positive bacteria, with subsequent PCR and GenBank analyses resulted in 16S rRNA gene sequences that clustered the residual Lactobacillus strain with the Spanish HG794492 (acc. no.) Lactobacillus strain, AY681132 Lactobacillus vini and AB242320 Lactobacillus mobilis, with 75% 16S rRNA gene sequence similarity. In conclusion, findings in the study provided important operational information that was previously unknown about the residual bacterial strain in cucurbitacin phytonematicides.
Keywords
Actinomycete bacteria; Bacteriocins; Lactic acids; Lactobacillus species; Photosynthetic bacteria; Salivaricins
Abbrevations
EM: Effective Microorganisms; EDTA: Ethylenediamine Tetraacetic Acid; MRS: Magnetic Resonance Spectroscopy; LBC: Liquid-Based Cytology; ARB: Antibiotic Resistant Bacteria; RNA: Ribonucleic Acid; DNA: Deoxyribonucleic Acid; ATP: Adenosine Triphosphate.
Introduction
Post withdrawal of the fumigant chemical nematicides from the agrochemical markets, cucurbitacin phytonematicides, produced through fermentation using Effective Microorganisms (EM) had high efficacy in suppressing nematode population densities, especially the root-knot (Meloidogyne species) nematodes. The products became integral in alternative products intended to manage nematode population densities in various cropping systems [1]. Broadly, EM constituents include the unidentified lactic acid bacteria, yeast bacteria, photosynthetic bacteria, actinomycetes, along with certain minor fungi and mycoplasms [2]. Roles of EM constituents had been articulated, but with limited information on the species used as part of protecting the intellectual property of the stock EM [2]. In EM technology, local bacterial strains are used in formulation of stock EM solutions in order to protect indigenous strains from interference, where the introduced bacterial strains almost always displace local strains to extinction. Among other roles, EM constituents have the capability to produce antimicrobial chemicals that sterilize phytonematicides, ensuring that the products remain aseptic for extended periods.
Cucurbitacin phytonematicides, as alternatives to fumigant chemical nematicides, are EM-extracted from dried fruits of wild cucumber (Cucumis myriocarpus) and wild watermelon (Cucumis africanus) [1]. The products from the respective plants are available as Nemarioc-AL and Nemafric-BL phytonematicides, with A and B being active ingredients cucurbitacin A (C32H46Os) and B (C32H48O8), respectively, whereas L is indicative that the product is in liquid formulation. Cucurbitacins are highly oxygenated triterpenoids with tetracyclic cucurbitane nucleus skeletons: (the 19-(10 → β)-abeo-10 α-lanost-5-ene [3,4]. Although cucurbitacins have a common nucleus skeleton, the primary biosynthesized cucurbitacins in plant organs are cucurbitacin B and E, each with an acetyl function at C-25 [5]. All other known cucurbitacins, at least 20, are produced from the two primary cucurbitacins either through hydrogenation by cucurbitacin 23–reductase, deacetylation by cucurbitacin acetylesterases, hydroxylation, dehydrogenation or isomerization chemical reactions [5-8]. Generally, cucurbitacins differ from one another from hydroxylation at C-2, C-3, C-19 and C-24, along with the existence of a double chemical bonds between C-1 and C-2 or between C-23 and C-24, the acetylation of C-25 hydroxyl groups and the presence of a ketone function at C-3 [3,4,8]. Such chemical arrangements confer a wide array of biological activities to cucurbitacins. Pharmacological studies had since shown that cucurbitacins have the potential to treat various human diseases [3,4,9-11]. In traditional medicine, plant organs with cucurbitacins had been used for treatment of various human and animal diseases [9], including the deworming of canines [12].
During EM fermentation, unidentified lactic acid bacteria are responsible for degradation of cellulose (C6H10O5)n, a polysaccharide comprising linear chains of several hundreds to thousands of β (1 → 4)-linked D-glucose units, which constitute pentose structures, cemented together by lignin [13,14]. The resultant structure being cellulose, is the toughest material in plant cell walls [15]. The endproducts of degradation of cellulose are lactic acid molecules, with capabilities to reduce pH of plant-based fermented products [2]. Yeast bacteria, is responsible for anaerobic breakdown of glucose (glycolysis) to pyruvic acid [16], which also lowers pH to acidic medium, contributing to decreasing pH from 7.0 to 3.7 at 14 days after initiating the fermentation process. Such acidic conditions stimulate the multiplication of bacteria, but inhibit growth of fungi [2]. Additionally, yeast bacteria release antimicrobial chemicals that add to the bio-sterilization of phytonematicides by preventing growth of pathogenic microbes that include Penicillium simplicissimum, the fungus associated with post-harvest fruit-rotting of Cucumis species [9]. Although this fungus has plant growth stimulating attributes [17], in its pure form it is not desirable for use as an agricultural input since it produces tremorgenic mycotoxins that might contaminate produce [18]. EM-fermented plant materials release copious quantities of sulfur (S) as Hydrogen Sulfured (H2S), used by photosynthetic bacteria, which is a constituent of EM, that reduces NAD to NADH by H+ ion from H2S, as illustrated during Photosystem II of photosynthesis, where H+ from H2O molecule is used in the reduction process [19]. Sulfur is promptly oxidized to a lethal gas, SO42–, which is quickly reduced to sulfuric acid (H2SO4) by 2H+ from water (H2O) molecules–a strongly corrosive acid. The oxygen-tolerant actinomycete bacteria have the ability to release chitinase that hydrolyzes chitin in exoskeletons of insects, insect eggs, nematode eggs and fungal mycelia [9]. Apparently, none of the EM constituents releases reductase enzymes that biodegrade cucurbitacins [20]. The residual bacteria in EM constituents post fermentation of cucurbitacin phytonematicides had not been documented. The objective of this study, therefore, was to use phenotypic traits and molecular approaches to investigate the residual bacteria in stored cucurbitacin phytonematicides, along with its potential role in retaining the products aseptic, along with their potential survival mode.
Materials and Methods
Phenotypic trait tests
Unidentified residual bacterial strains from well-mixed Nemafric- BL phytonematicide samples from six 50 ml-plastic containers were collected at 35 weeks after storage for assays. The strains were mixed and further grown in modified MRS (mMRS) broth comprising MRS (Scharlab) supplemented with L-cysteine hydrochloride at 0.5 g L-1 at 28ºC for 4 days [21]. The specimens were placed in air-sealed plastic bags and stored in 20% (v/v) glycerol at -80ºC for two weeks prior to phenotypic characterization tests using standard analytical procedures [20-28]. Width and length of rod-shaped strains were measured using ZEISS microscope equipped with AmScope digital measuring software. Specimens were subjected to the Heimbrook staining method [24], fixed in oil and assessed for the presence of flagella under the compound microscope at 100 X magnification.
DNA extraction, PCR and phylogenetic analysis
The DNA extraction was accomplished using protocols outlined in ZymoBIOMICSTM DNA Miniprep Kit for Gram-positive bacteria in Liquid-Based Cytology (LBC) specimens from a 35-week-stored Nemafric-BL phytonematicide [29-31]. Briefly, samples of LBC specimens were dispensed into four 2 ML sterile collection tubes (volume 500 μL) to form a sample size of 20 DNA extractions, with negative control comprising ThinPrep preservation solution as blank extraction [31].
16rRNA marker gene sequencing
Control and extracted DNA was sent to Inqaba Biotechnical Industries (Pty) Ltd (Pretoria, South Africa) for amplification and sequencing of the 16S rRNA gene on an IlluminaR MiSeqTM sequencing platform. Overlapping paired-end reads were assembled into complete amplicon sequences. Amplicon sequences were profiled with Qiime using 16S rRNA gene database (gg-13-8) [32,33]. PCR was conducted with 10-15 ng DNA template, 12.5 μl of 2X PCR Master Mix Red (Promega, USA) in 1 μl of each primer (10 pmol μl-1) and with dd H2O added for a final volume at 30 μl. Amplification of 16S rRNA gene was processed using an Eppendorf Master Cycler Gradient (Eppendorf, Hamburg, Germany), using the stepwise procedure of initial denaturation for 3 min at 94°C, followed by 37 cycles of denaturation for 45 s at 94°C, and then 50°C annealing temperature, with extension for 45 s to 1 min at 72°C. The final extension step of 6 min at 72°C was followed by a temperature on-hold at 4°C. After DNA amplification, 4 μl eluted DNA was loaded on a 1% agarose gel in TBE buffer (40 mM Tris, 40 mM boric acid and one mM EDTA) for assessing DNA bands after staining with RedGel and photographing on a UV transilluminator for visualization.
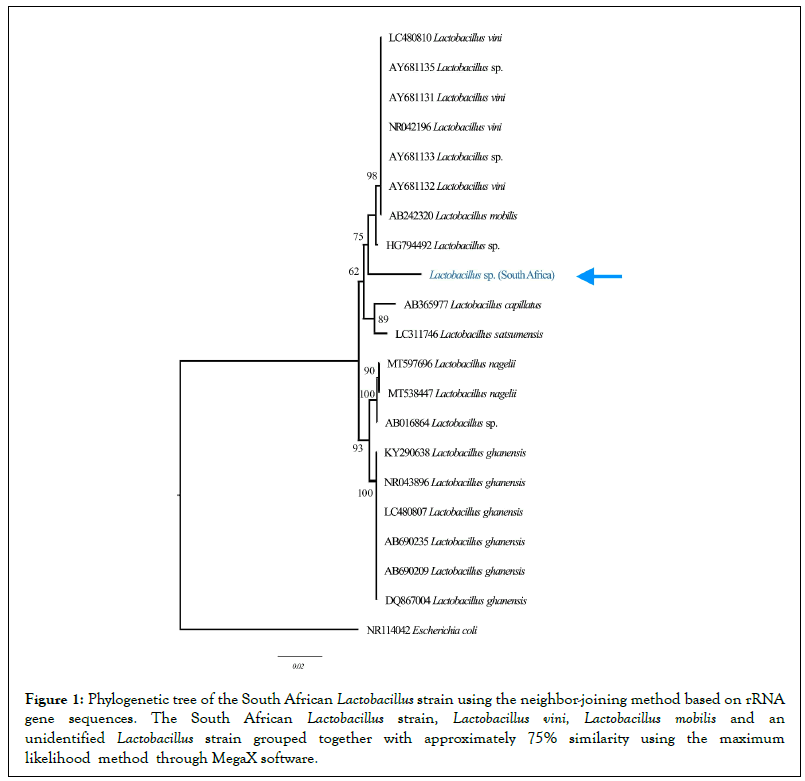
GenBank accession numbers of 16S rRNA gene sequences of 500 bp corresponding to the phenotypically-identified South African Lactobacillus strain were conducted using the Mega-X software [34]. The data subsets were employed using the corresponding ARB program package [35]. Sequence similarity values were calculated by comparing nucleotides at the corresponding positions, using three alternative tree methods, namely, neighbor-joining, maximumparsimony and maximum-likelihood [36]. The tree in Figure 1 was constructed using the neighbor-joining method based on rRNA gene sequences [37], with NR114042 Escherichia coli used as an outgroup for comparison.
Results and Discussion
Phenotypic traits of residual strain
Width and length of the rod-shaped strain were 0.49-0.82 μm and 1.36-2.8 μm, respectively, with mean value of 0.72 × 2.24 μm. Cells were non-spore forming, occurring singly, with occasional pairs of 2-5. No flagella were detected under the Heimbrook staining method, but most cells on wet mounts of mMRS appeared motile. The facultative anaerobic test strain was Gram-positive and catalasenegative. Using the L-lactic kit and HPLC [23], the strain formed DL-lactate from glucose, without release of CO2 or inducing fermentation of gluconate, and thus the strain was categorized as having homofermentative process [36]. Additionally, the strain had transaldolase and transketolase activities, suggesting the capability to use pentose sugars from cellulose through the pentose phosphate pathway to yield lactates as end-products, which is a distinct feature in 6-phosphogluconate pathway that is commonly used by fermentative facultative bacterial strains [26]. Additionally, the test strain grew at 25ºC, 37ºC and 45ºC, but never at 5ºC or 15ºC, as well as at pH 3.7, 4.5 and 8.0 in media adjusted with either 5% or 10% NaCl (w/v). Additionally, the bacterial strain produced neither exopolysaccharides from sucrose nor any gas from glucose. Generally, the described phenotypic features characterize Lactobacillus strains [35]. Therefore, we referred to the test strain that occurred as the residual bacterial strain post EM-fermentation of Nemafric-BL phytonematicide as the South African Lactobacillus strain.
Phylogenetic analysis
The phylogenetic tree was constructed using maximum-likelihood analysis of 16S rRNA gene. The latter clustered the South African Lactobacillus strain with the Spanish HG794492 (acc. No.) Lactobacillus strain, AY681132 Lactobacillus vini and AB242320 Lactobacillus mobilis, with 75% 16S rRNA gene sequence similarity (Figure 1). Comparison of 16S rRNA gene sequence of the South African Lactobacillus strain with corresponding B242320 Lactobacillus mobilis from the GenBank database showed 87% similarity with 151 nucleotide differences. Additionally, comparison of 16S rRNA gene sequence of the South African Lactobacillus strain with AY681132 Lactobacillus vini had 87% similarity with 151 nucleotide differences [38]. Generally, Lactobacillus vini bacterial strains are Gram-positive, motile, facultative anaerobic rods, catalase-negative, with the ability to biodegrade cellulose by attacking pentose sugars through the homofermentative process that produces lactic acid as end-products [35] as confirmed in our phenotypic tests.
Figure 1: Phylogenetic tree of the South African Lactobacillus strain using the neighbor-joining method based on rRNA gene sequences. The South African Lactobacillus strain, Lactobacillus vini, Lactobacillus mobilis and an unidentified Lactobacillus strain grouped together with approximately 75% similarity using the maximum likelihood method through MegaX software.
The high similarity of the South African Lactobacillus strain with Lactobacillus vini in EM provided cogent explanation on why lactic acid bacteria in EM was viewed as being capable of biodegrading cellulose [2]. Generally, Lactobacillus vini bacterial strains are dominant in South African soils since such strains are widespread as contaminants in winemaking and ethanol fermentation industries. Similarly, 16S rRNA nucleotide sequence BLAST of the South African Lactobacillus strain had a similarity from 85% to 87% 16S rRNA gene sequence with those of other Lactobacillus strains in the GenBank. The high similarities on phylogenetic tree of the South African Lactobacillus strain with other globally-existing Lactobacillus strains, confirmed other observations using maximum likelihood methods through MegaX software, where results showed that the genus Lactobacillus was a highly diversified genus [39]. Due to the closely relatedness of the South to other Lactobacillus strains, the South African-produced EM-fermented phytonematicides could be used in various countries without affecting the indigenous Lactobacillus species in importing countries.
Sustainability features of Lactobacillus strains in phytonematicides
Phenotypic and genomic outcomes of the current study showed that the residual bacteria in EM-fermented phytonematicides were Lactobacillus strains, where glucose molecules are metabolized through the phosphoketolase pathway, which comprise either the homolactic or heterolactic fermentative process [40]. Briefly, the two fermentative processes are summarized as follows:
• Homolactic fermentative process: glucose+2ADP+Pi 2lactic acids+2ATP
• Heterolactic fermentative process: glucose+ADP+Pi lactic acids+ethanol+CO2+ATP
The phenotypic outcomes suggested that the South African Lactobacillus strain was proficient in homolactic fermentative process, which is more efficient than the other since one glucose molecule is metabolized to two lactic acids and two ATP molecules. In both processes, Pi is phosphorus derived from the substrate [19]. Another advantage of the South African Lactobacillus strain that it does not release CO2 as end-product, which would result in accumulation of gases and the subsequent bulging of containers. Further, the released lactic acids and hydrogen peroxide (H2Os) in homolactic fermentation process, further suppress other anaerobic bacteria except for Lactobacillus strains, which are acid and oxygen-tolerant [40,41]. Due to the latter attribute, when opening containers during operations, there is no need to take precautionary measures related to exposure oxygen. Lactobacillus strains also produce copious quantities of bacteriocins, salivaricins and sodium butyrate, which are antimicrobial chemicals to various pathogenic microbes [42]. The existence of these antimicrobial chemicals provide an explanation why cucurbitacin phytonematicides remain highly aseptic for extended periods, regardless of repeated opening of containers during operations [43].
Conclusion
Use of cucurbitacin phytonematicides is one of the most prominent alternative strategies for managing nematode population densities after the withdrawal of fumigant chemical nematicides from the agrochemical markets. Effective microorganisms used in fermentation of phytonematicides comprise unidentified strains of lactic acid bacteria, yeast bacteria, photosynthetic bacteria, actinomycetes bacteria and other minor entities such as mycoplasms and fungi, with each being country-specific. Isolation, phenotypic testing and next generation sequencing were performed on residual bacteria at 35 weeks post fermentation of Nemafric- BL phytonematicide. The unidentified lactic-acid bacteria, phenotypically tagged the South African Lactobacillus strain, was clustered with five Lactobacillus strains, with 75% 16S rRNA gene sequence similarity. Similarity with Lactobacillus vini was of interest since the strain was previously used to explain how lactic acid bacteria in EM biodegrade cellulose, along with providing evidence on how cucurbitacin phytonematicides remain aseptic for extended periods.
Acknowledgement
This work is based on the research supported wholly by the National Research Foundation of South Africa (Grant Number 136124).
REFERENCES
- Mashela PW, Waele DD, Dube Z, Khosa MC, Pofu KM, Tefu G, et al. Alternative nematode management strategies. InNematology in South Africa: a view from the 21st century. 2017; 151-181.
- Higa T, Parr JF. Beneficial and effective microorganisms for a sustainable agriculture and environment. Atami: International Nature Farming Research Center. 1994 . [Crossref],
[Google scholar], [Indexed]
- Chen JC, Chiu MH, Nie RL, Cordell GA, Qiu SX. Cucurbitacins and cucurbitane glycosides: Structures and biological activities. Natural Product Reports. 2005; 22(3): 386-399.
[Crossref], [Google scholar], [Indexed]
- Chen CH, Kuo TC, Yang MH, Chien TY, Chu MJ, Huang LC, et al. Identification of cucurbitacins and assembly of a draft genome for Aquilaria agallocha. BMC genomics. 2014; 15(1): 1-1.
[Crossref], [Google scholar], [Indexed]
- Gry J. Cucurbitacins in plant food. Nordic Council of Ministers. 2006.
- Dirr HW, Schabort JC, Weitz C. Cucurbitacin Δ23-reductase from the fruit of Cucurbita maxima var. Green Hubbard. Physicochemical and fluorescence properties and enzyme-ligand interactions. Biochemical journal. 1986; 233(3): 649-653.
[Crossref], [Google scholar], [Indexed]
- Schabort JC, Teijema HL. The role of cucurbitacin Δ23-reductase in the breakdown pathway of toxic bitter principles in Cucurbita maxima. Phytochemistry. 1968; 7(12): 2107-2110.
[Crossref], [Google scholar]
- Zhou Y, Ma Y, Zeng J, Duan L, Xue X, Wang H, et al. Convergence and divergence of bitterness biosynthesis and regulation in Cucurbitaceae. Nature plants. 2016; 2(12): 1-8.
[Crossref], [Google scholar], [Indexed]
- Mphahlele RR, Mashela PW, Pofu KM. Post-harvest fruit decay-inducing pathogen in medicinally important Cucumis species indigenous to South Africa. Afr J Agric. Res. 2012; 7(26): 3786-3791.
[Crossref], [Google scholar]
- Abbas S, Vincourt JB, Habib L, Netter P, Greige-Gerges H, Magdalou J. The cucurbitacins E, D and I: investigation of their cytotoxicity toward human chondrosarcoma SW 1353 cell line and their biotransformation in man liver. Toxicology letters. 2013; 216(2-3): 189-199.
[Crossref], [Google scholar], [Indexed]
- Mir SA, Mukherjee S, Makar S, Pal G. Cucurbitacins a vibrant triterpenoid: A review on its anticancer property. PharmaTutor. 2019; 7(2):43-54.
[Crossref], [Google scholar]
- Mashela PW, Dirk De Waele D, Pofu KM. Use of indigenous Cucumis technologies as alternative to synthetic nematicides in management of root-knot nematodes in low-input agricultural farming systems: A review. Scientific Research and Essays. 2011; 6(33): 6762-6768.
[Crossref], [Google scholar]
- Crawford RL. Lignin biodegradation and transformation.
- Updegraff DM. Semimicro determination of cellulose inbiological materials. Anal. Biochem. 1969; 32(3): 420-424.
[Crossref], [Google scholar], [Indexed]
- de Vries L, Guevara-Rozo S, Cho M, Liu LY, Renneckar S, Mansfield SD. Tailoring renewable materials via plant biotechnology. Biotechnol Biofuels. 2021; 14(1): 1-33.
[Crossref], [Google scholar], [Indexed]
- Stetter KO. History of discovery of the first hyperthermophiles. Extremophiles. 2006; 10(5): 357-362.
[Crossref], [Google scholar]
- Hossain MM, Sultana F, Kubota M, Koyama H, Hyakumachi M. The plant growth-promoting fungus Penicillium simplicissimum GP17-2 induces resistance in Arabidopsis thaliana by activation of multiple defense signals. Plant and cell physiology. 2007; 48(12): 1724-1736.
[Crossref], [Google scholar], [Indexed]
- Pitt JI. Penicillium crustosum and P. simplicissimum, the correct names for two common species producing tremorgenic mycotoxins. Mycologia. 1979; 71(6): 1166-1177.
[Crossref], [Google scholar]
- Campbell NA, Reece JB, Mitchell LG. Biology Benjamin/Cummings. Menlo Park, CAMulligan E. 1999: 907-910.
- Ellis EM. Microbial aldo-keto reductases. FEMS microbiology letters. 2002; 216(2): 123-131.
[Crossref], [Google scholar], [Indexed]
- Rodas AM, Chenoll E, Macian MC, Ferrer S, Pardo I, Aznar R. Lactobacillus vini sp. nov., a wine lactic acid bacterium homofermentative for pentoses. Int J Syst Evol Microbiol. 2006; 56(3): 513-517.
[Crossref], [Google scholar], [Indexed]
- 22. Barre P. Identification of thermobacteria and homofermentative, thermophilic, pentose‐utilizing lactobacilli from high temperature fermenting grape musts. J Appl Microbiol 1978; 44(1): 125-129.
[Crossref], [Google scholar]
- Frayne RF. Direct analysis of the major organic components in grape must and wine using high performance liquid chromatography. Am J Enol Vitic 1986; 37(4): 281-287.
- Heimbrook ME, Wang WL, Campbell GA. Staining bacterial flagella easily. J Clin Microbiol. 1989; 27(11): 2612-2615.
[Crossref], [Google scholar], [Indexed]
- Kandler O. Carbohydrate metabolism in lactic acid bacteria. 1983; 49(3): 209-224.
[Crossref], [Google scholar], [Indexed]
- Nihira T, Saito Y, Nishimoto M, Kitaoka M, Igarashi K, Ohtsubo KI, et al. Discovery of cellobionic acid phosphorylase in cellulolytic bacteria and fungi. FEBS letters. 2013; 587(21): 3556-3561.
[Crossref], [Google scholar], [Indexed]
- Rodas AM, Ferrer S, Pardo I. Polyphasic study of wine Lactobacillus strains: Taxonomic implications. Int J Syst Evol. Microbiol. 2005; 55(1): 197-207.
[Crossref], [Google scholar], [Indexed]
- Picataggio SK, Zhang M, Franden MA, Mc Millan JD, Finkelstein M, et al. Recombinant lactobacillus for fermentation of xylose to lactic acid and lactate. 1998.
- Ravilla R, Coleman HN, Chow CE, Chan L, Fuhrman BJ, Greenfield WW, et al. Cervical microbiome and response to a human papillomavirus therapeutic vaccine for treating high-grade cervical squamous intraepithelial lesion. Integrative cancer therapies. 2019; 18: 1534735419893063.
[Crossref], [Google scholar], [Indexed]
- Zymo Research ZymoBiomicsTM DNA Miniprep Kit.
- Shibata T, Nakagawa M, Coleman HN, Owens SM, Greenfield WW, Sasagawa T, et al. Evaluation of DNA extraction protocols from liquid-based cytology specimens for studying cervical microbiota. Plos one. 2021; 16(8): e0237556.
[Crossref], [Google scholar], [Indexed]
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019; 37(8): 852-857.
[Crossref], [Google scholar], [Indexed]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the national academy of sciences. 2011; 108: 4516-4522.
[Crossref], [Google scholar], [Indexed]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004; 32(4):1363-1371.
[Crossref], [Google scholar], [Indexed]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018; 35(6): 1547.
[Crossref], [Google scholar], [Indexed]
- Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, Neumaier J, et al. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis. 1998; 19(4): 554-568.
[Crossref], [Google scholar], [Indexed]
- Collins MD, Rodrigues U, Ash C, Aguirre M, Farrow JA, Martinez-Murcia A, et al. Phylogenetic analysis of the genus Lactobacillus and related lactic acid bacteria as determined by reverse transcriptase sequencing of 16S rRNA. FEMS Microbiology letters. 1991; 77(1): 5-12.
[Crossref], [Google scholar]
- Stackebrandt E, GOEBEL BM. Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Evol Microbiol. 1994; 44(4): 846-849.
[Crossref], [Google scholar]
- Rodas AM, Chenoll E, Macian MC, Ferrer S, Pardo I, Aznar R. Lactobacillus vini sp. nov., a wine lactic acid bacterium homofermentative for pentoses. Int J Syst Evol Microbiol. 2006; 56(3): 513-517.
[Crossref], [Google scholar], [Indexed]
- Claesson MJ, van Sinderen D, O'Toole PW. Lactobacillus phylogenomics–towards a reclassification of the genus. Int J Syst Evol Microbiol. 2008; 58(12): 2945-2954.
[Crossref], [Google scholar], [Indexed]
- Vidali M. Bioremediation: An overview. Pure Appl Chem. 2001; 73(7): 1163-1172.
[Crossref], [Google scholar]
- Barbour A, Wescombe P, Smith L. Evolution of lantibiotic salivaricins: new weapons to fight infectious diseases. Trends Microbiol. 2020; 28(7): 578-593.
[Crossref], [Google scholar], [Indexed]
- Mashela PW, Shokoohi E, Pofu KM. Shelf-life in cucurbitacin-containing phytonematicides: Non-conformity to Arrhenius model. Plos One. 2020; 15(2): e0227959.
[Crossref], [Google scholar], [Indexed]
Citation: Mashela PW, Pofu KM (2022) Phenotypic And Phylogenetic Analyses of Residual Bacteria in Effective Microorganism-Fermented Cucurbitacin Phytonematicide. J Plant Pathol Microbiol. 13:606.
Copyright: © 2022 Mashela PW, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License,which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.