Indexed In
- Academic Journals Database
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- China National Knowledge Infrastructure (CNKI)
- Ulrich's Periodicals Directory
- Electronic Journals Library
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Commentary Article - (2023) Volume 14, Issue 4
Peptide Epitopes as Biomarkers in Allergology
Szardenings Michael1*, Kern Karolin1 and Treudler Regina22Department of Dermatology, Venereology and Allergology, University Leipzig Medical Faculty, Leipzig, Germany
Received: 16-Jun-2023, Manuscript No. JAT-23-21894 ; Editor assigned: 19-Jun-2023, Pre QC No. JAT-23-21894 (PQ); Reviewed: 03-Jul-2023, QC No. JAT-23-21894 ; Revised: 10-Jul-2023, Manuscript No. JAT-23-21894 (R); Published: 18-Jul-2023, DOI: 10.35248/2156-6121.14.352
Abbreviations
SPT: Skin Prick Test; EoT: End of Treatment; LOAELo/s: Lowest Observed Adverse Effect Level (Objective/Subjective signs) in food challenge.
Description
Allergies count for one of the most frequent diseases worldwide and include a broad spectrum of chronic conditions such as allergic rhinitis, allergic asthma or food allergy [1]. Quality of life is frequently affected and the cornerstone of clinical care consists in allergen identification. When allergen avoidance cannot be realized, symptomatic and, if available, allergen-specific therapy is indicated. Still there exist multiple limitations in allergy diagnostic and therapy [1].
In IgE-mediated allergies, e.g. in pollen or food allergy, skin prick tests and determination of specific IgE values are standard diagnostic measures. The recent introduction of molecular allergy diagnostics based on individual protein molecules of the allergen was helpful in detecting relevant IgE sensitization and sometimes even allows to predict the risk of severity of clinical reactions.
There are several limitations, though:
• The wide range of possible allergens cannot be covered by commercially available test systems.
• Similarity of sequence and structure of in particular plant allergens lead to high rates of false positive results with current protein-based assays [2,3].
Several IgE-mediated allergies, for example: against pollen, house dust mites and hymenoptera can effectively be treated by Allergen-specific Immunotherapy (AIT). This requires proper diagnosis of the allergy but biomarkers are still needed to stratify patients and identify those who will benefit from AIT.
To date, the amount of antibodies binding individual allergenic proteins or skin prick test results are not able to predict neither the course nor the outcome of AIT [4]. The current knowledge regarding the epitope diversity and specificity of allergen-specific antibodies induced during AIT is limited [5-7].
Optimized peptide phage display methods have been used for the identification of peanut [8,9], and soya epitopes recognized by sensitized patient sera [6,9-11]. Remarkably, an IgG wheat epitope was able to discriminate between non-sensitized and sensitized wheat patients and was identified by using an allergen peptide phage display library [12]. We reported the identification and validation of a large number of soya epitopes and mimotopes (i.e. peptides that mimic conformational epitopes) by applying a unique peptide phage display platform that involves the statistical analysis of enriched allergen-motifs. This has the advantage to directly identify binding peptide epitopes even with structural constraints from a naïve peptide library.
Recently, we presented a paper on the use of peptide epitopes as potential epitope-based markers for monitoring and/or predicting AIT efficacy [13]. We included sera from patients having been treated within a clinical trial by AIT applying a recombinant folded variant of the main birch pollen allergen Bet v1 [14]. Clinical relevance of epitopes was evaluated by comparing these measurements to a number of treatment outcome measures recorded during double-blind placebocontrolled food challenges at start and end of AIT [13].
This study was only made possible by applying a novel method to analyse the results from binding a naïve peptide library on bacteriophage to the entire Ig-pool of the patient sera and analyzing the data of bound sequences without repeated selection rounds. In addition a peptide micro-array of 320 peptides resembling epitopes of 15 food allergens also comprising multiple epitopes from PR10 variants was used. Altogether these investigations used only about 100 μl serum remaining form the earlier BASALIT study. Peptide arrays were generated in another large study involving more than 500 patients, which is presently under evaluation for research and diagnostics applications (Fraunhofer Project Food Allergen). In particular the data analysis can be carried out in silico for protein sequences of multiple antigens [15], which otherwise would require large quantities of serum screening multiple peptide arrays generated from peptide fragments of the individual proteins. We found already earlier that the phage method was superior in the direct comparison to peptide arrays [10]. It allows the selection of peptides matching multiple sera and using the naïve library peptides are identified that contain structurally relevant amino acids, for example- cysteine, adjacent to the binding motif. Such peptides can have a much-improved affinity and selectivity and allow individual measurements of IgE and IgG. The recent study on using such epitopes resembling PR10 epitopes would not have been possible without such rather short and structurally confined sequences.
Binding of IgE and IgG to individual peptide epitopes can be a potential indicator of soya sensitization at start of treatment and distinct IgG-epitopes may help to predict and monitor clinical effects of AIT. With respect to the overall expected cross reactivity of PR10 antibodies, selected peptide epitopes are probably the best tool for proper diagnosis and prognosis not only in food allergies, but also other immune disease.
In summary, reactivity to mainly IgG-epitopes may help to predict clinical effects of AIT on soya and possibly also in other types of IgE-mediated allergy. Our measurements show that also IgG epitopes can be biomarkers of multiple clinical measures in allergies (Figure 1).
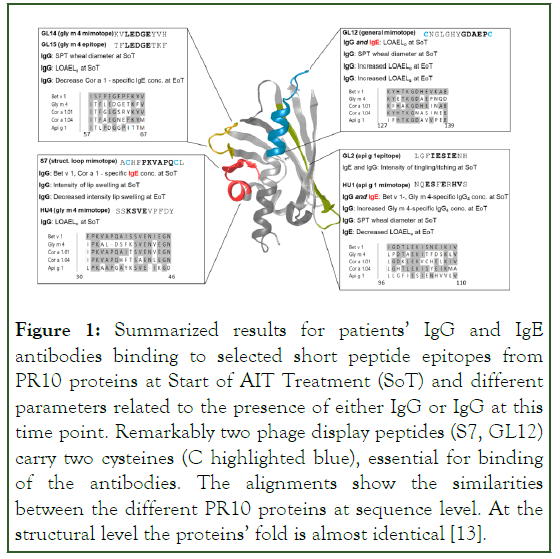
Figure 1: Summarized results for patients’ IgG and IgE antibodies binding to selected short peptide epitopes from PR10 proteins at Start of AIT Treatment (SoT) and different parameters related to the presence of either IgG or IgG at this time point. Remarkably two phage display peptides (S7, GL12) carry two cysteines (C highlighted blue), essential for binding of the antibodies. The alignments show the similarities between the different PR10 proteins at sequence level. At the structural level the proteins’ fold is almost identical [13].
References
- Treudler R, Simon JC. Developments and perspectives in allergology. J Dtsch Dermatol Ges. 2023;21(4):399-403.
[Crossref] [Google Scholar] [PubMed]
- Treudler R, Simon JC. Overview of component resolved diagnostics. Curr Allergy Asthma Rep. 2013;13:110-117.
[Crossref] [Google Scholar] [PubMed]
- Ansotegui IJ, Melioli G, Canonica GW, Caraballo L, Villa E, Ebisawa M, et al. IgE allergy diagnostics and other relevant tests in allergy, a World Allergy Organization position paper. World Allergy Organ J. 2020;13(2):100080.
[Crossref] [Google Scholar] [PubMed]
- Treudler R, Klimek L. Allergen immunotherapy for oral allergy syndrome: What is the evidence for efficacy?. Aller J Int. 2019;28:50-56.
- Eiwegger T, Hung L, San Diego KE, O'Mahony L, Upton J. Recent developments and highlights in food allergy. Allergy. 2019;74(12):2355-2367.
[Crossref] [Google Scholar] [PubMed]
- Ehlers AM, Blankestijn MA, Knulst AC, Klinge M, Otten HG. Can alternative epitope mapping approaches increase the impact of B‐cell epitopes in food allergy diagnostics?. Clin Exp Allergy. 2019;49(1):17-26.
[Crossref] [Google Scholar] [PubMed]
- Pomés A, Mueller GA, Chruszcz M. Structural aspects of the allergen-antibody interaction. Front Immunol. 2020;11:2067.
[Crossref] [Google Scholar] [PubMed]
- Christiansen A, Kringelum JV, Hansen CS, Bøgh KL, Sullivan E, Patel J, et al. High-throughput sequencing enhanced phage display enables the identification of patient-specific epitope motifs in serum. Sci Rep. 2015;5(1):12913.
[Crossref] [Google Scholar] [PubMed]
- Chen X, Dreskin SC. Application of phage peptide display technology for the study of food allergen epitopes. Mol Nutr Food Res. 2017;61(6).
- Kern K, Havenith H, Delaroque N, Rautenberger P, Lehmann J. The immunome of soy bean allergy: Comprehensive identification and characterization of epitopes. Clin Exp Allergy. 2019;49(2):239-51
[Crossref] [Google Scholar] [PubMed]
- Havenith H, Kern K, Rautenberger P, Spiegel H, Szardenings M, Ueberham E, et al. Combination of two epitope identification techniques enables the rational design of soy allergen Gly m 4 mutants. Biotechnol J. 2017;12(2):1600441.
[Crossref] [Google Scholar] [PubMed]
- Monaco DR, Sie BM, Nirschl TR, Knight AC, Sampson HA, Nowak-Wegrzyn A, et al. Profiling serum antibodies with a pan allergen phage library identifies key wheat allergy epitopes. Nat Commun. 2021;12(1):379.
[Crossref] [Google Scholar] [PubMed]
- Caballero LR, Treudler R, Delaroque N, Simon JC, Kern K, Szardenings M. Peptide epitopes as biomarkers of soya sensitization in rBet v 1 immunotherapy of birch-related soya allergy. Clin Exp Allergy. 2023;53(3):316–26.
[Crossref] [Google Scholar] [PubMed]
- Treudler R, Franke A, Schmiedeknecht A, Ballmer‐Weber B, Worm M, Werfel T, et al. BASALIT trial: double‐blind placebo‐controlled allergen immunotherapy with rB et v 1‐FV in birch‐related soya allergy. Allergy. 2017;72(8):1243-1253.
[Crossref] [Google Scholar] [PubMed]
- Ramírez Caballero L, Kny C, Treudler R, Simon JC, Kern K, Jappe U, et al. Identification of Seasonal Variations of Antibodies against PR-10-Specific Epitopes Can Be Improved Using Peptide-Phage Display. Int Arch Allergy Immunol. 2020;181(12):919-25.
[Crossref] [Google Scholar] [PubMed]
Citation: Michael S, Karolin K, Regina T (2023) Peptide Epitopes as Biomarkers in Allergology. J Allergy Ther. 14:352.
Copyright: © 2023 Michael S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.