Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- China National Knowledge Infrastructure (CNKI)
- Ulrich's Periodicals Directory
- RefSeek
- Hamdard University
- EBSCO A-Z
- Directory of Abstract Indexing for Journals
- OCLC- WorldCat
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2016) Volume 6, Issue 4
Osteogenic Differentiation of Human Umbilical Cord Peri-vascular Cells using Low Intensity Pulsed Ultrasound
Abstract
The objective of this experiment was to explore the possible effect of low intensity pulsed ultrasound (LIPUS) on the osteogenic differentiation of harvested passage-4 Human umbilical cord peri-vascular cells (HUCPV-Cs). HUCPV-Cs were divided into two groups: a treatment group that received LIPUS for 10 minutes for 1, 7 and 14 days and a control group that received a sham treatment utilizing osteogenic media. The results demonstrated nonsignificant differences in cell count, ALP, DNA content, and CD90. Statistically significant expression of OPN and PCNA was observed on day 14 in the LIPUS treated group. Nucleostemin expression in the LIPUS-treated group was nonsignificant on days 1 and 7. However, a selective increase in the osteogenic markers was observed in the LIPUS treated group on day 7 for ALP and OCN and on day 14 for OPN. Future experiments are required to explore the possible effects of different application times and/or techniques of LIPUS on the behaviour of HUCPV-Cs.
Keywords: Low Intensity Pulse Ultrasound; Stem cells; Human Umbilical Cord Perivascular Cells; Osteogenic differentiation
Introduction
Mesenchymal stem cells represent a promising future for medicine [1,2]. They introduce a new tool for clinical concepts that support cellular therapy [1,2]. Obtaining MSCs from bone marrow is an invasive procedure [1,3]. Frequency, differentiation potential, and life span of bone marrow-MSCs decrease with age [4-6]. Therefore, many researches have been investigated an alternative source for mesenchymal stem cells in order to find a cure for many ailments [6]. It has been shown that cryopreserved umbilical cord blood from unrelated donors is a safe source of transplantable hematopoietic stem cells for clinical transplantation [6]. They reported that those cells have high rate of engraftment and low rate of grade III- IV acute graft versus host disease (GVHD) and even in recipients of human leukocyte antigens (HLA) unrelated grafts are remarkable [6]. Umbilical cord cells have been progressively used as an alternative source for hematopoietic stem cells (HSC) for allogenic stem cell transplants [3-8]. It has been reported that MSCs from bone marrow, umbilical cord blood and adipose tissue can achieve a success in stem cell therapy [9]. Their clinical application may be based on their capacity of differentiation, but much more on their frequency, and expansion potential [9]. However, the lack of common standards for initial cell preparation remains an obstacle for standardization of research methodology and the clinical application of umbilical cord-MSCs [10,11].
“Mesenchymal stem cells derived from the umbilical cord vein are functionally similar to bone marrow MSCs” [12]. Isolation of umbilical cord MSCs (UCMSCs) is less invasive than bone marrow derivations, and because of the fetal origin of UCMSCs, their proliferative and differentiation potential provide an excellent resource [12]. In a comparative study, it has been documented that human umbilical cord perivascular cells (HUCPV-Cs) have higher capacity of differentiation and proliferation than bone marrow MSCs [13]. In addition, HUCPVCs were shown to have a faster rate of osteogenic differentiation compared to bone marrow MSCs [13]. Umbilical cord provides a pool of cells of vast abundance, and with the advantage of less donor site morbidity [14]. Gang et al. reported that umbilical cord blood derived cells (UCB-DCs) express high potential to differentiate into variety of mesenchymal linages cells [15]. He also claimed that UCBDCs is an excellent substitute source for human-MSCs [15]. Human umbilical cord stromal cells express almost the same characteristics of mesenchymal stem cells [16,17]. It has been reported that the umbilical cord stromal cells show high capability to differentiate into osteogenic, adipogenic, cardiomyogenic, and chondrogenic cell types [16]. A new technique for harvesting, culturing, and osteogenic differentiation of HUCPV-Cs has been reported [18]. Ultimately, the blood that remains inside the human umbilical cord is usually considered a valid source of hematopoietic stem cells [19,20]. Furthermore, Kim et al. reported that umbilical cord blood is an excellent source of profound mesenchymal progenitor cells (MPCs) characterized by the capacity for self-renewal and differentiation into multiple lineages which make them comparable to the same cells (MPCs) from different origin [21]. Goodwin et al. proven that umbilical cord blood cells are a potential source of cells for multiple organ cellular therapeutics [22]
Many reports have demonstrated that LIPUS enhances bone remodeling and bone formation as well as it decreases healing time [23-32]. Mechanical stresses have been reported to enhance activities of osteoclasts and osteoblasts leading to increase bone remodeling and bone regeneration, respectively [33]. Different forms of mechanical stress such as LIPUS have been clinically tested for their ability to enhance new bone formation [34].
Acceleration of fracture healing by LIPUS was attributed to the pressure waves that trigger a complex series of biochemical and molecular events at the cellular level [35]. An increase in alkaline phosphatase (ALP) activity was detected in human osteoblast cultures after continuous exposure to the low intensity pressure waves of LIPUS [36].
This study investigated whether LIPUS has a stimulatory effect on osteogenic differentiated HUCPV-Cs that can potentially increase the differentiation capacity of these cells during certain periods of time (10 minutes/day for 1,7 and 14 days). The influence of LIPUS was assessed using different methods including cell count, ALP assay, DNA assay, real-time PCR, and immunophenotyping of cells derived from HUCPV-Cs by flow-cytometry analysis.
Materials and Methods
This study has been approved by the Health Research Ethics Board at the University of Alberta, Edmonton, Canada (approval number 6431, 2006).
Cell culture
HUCPV-Cs were donated by Professor Joh E Davis at the University of Toronto at P0. HUCPV-Cs were obtained from patients undergoing full-term caesarean sections after obtaining standard patient’s consent and isolation of the cells were completed according to methods described by Sarugaser et al. [18].
HUCPV-Cs at passage 0 were thawed and seeded in T-75 cm2 tissue culture flasks (Sigma Aldrich). The cell cultured in osteogenic media included Dulbecco’s modified Eagle’s medium with low glucose (DMEM-LG) (GIBCO, Invitrogen) supplemented with 1% antibioticantimycotic (Sigma Aldrich), 15% fetal bovine serum (FBS), 5 mM β-glycerophosphate (Sigma Aldrich), 50 μg/ml L-ascorbic acid (Sigma Aldrich) [18], and 10–8 M dexamethasone (Sigma Aldrich). The cells were incubated at 37ºC in 5% CO2 and the initial cell density used was 3.6 × 106/ml. Ten days were implemented for the expansion of HUCPV cells until P3 and the media were changed every 2–3 days. The cells at P3 were harvested and trypsinized when their confluence reached 80% (4.2 × 106/ml) using 0.25% trypsin (GIBCO, Invitrogen). The cells at P4 were collected in 50 ml tubes, centrifuged, then plated into nine 6 well plates (Sigma Aldrich) at 2 × 104/ml. Four sets of “LIPUS” devices were obtained from SmileSonica Inc., Edmonton, Canada with 4 transducers placed below the wells and coupled to the well bases with standard ultrasound coupling gel transducers that was previously calibrated. A total of 27 wells were treated by these LIPUS devices for 10 min/day for 14 days where the ultrasound frequency, intensity, and duration were identical to that has been used for bone fracture repair experimentally and clinically [23-32]. Sham devices were used to treat the other 27 wells (Control Group) using the same transducers without turning on the machines. HUCPV-Cs were assessed for their differentiation capacity at day 1,7 and 14 consecutively. The ultrasound transducers generate 1.5-Mhz ultrasound waves of 200-μs bursts at an intensity of 30 mW/cm2 and pulse repetition frequency of 1 KHz. To maintain the consistency of electrical waveforms, the transducers were calibrated before and after applications using TDS1012C-EDU digital oscilloscope (Tektronix, Canada and an ultrasound powermeter (model UPM-DT-1AV from Ohmic Instruments, Easton, MD, USA). The incubator temperature was maintained at 37°C during the application of LIPUS.
Cell count
HUCPV-Cs were washed using PBS (GIBCO, Invitrogen, Burlington, ON, Canada) then trypsinized. Cells and medium were collected in 15 ml tubes and were spun for 6 minutes at 600 rpm (Treated group separated from Control group). The supernatant was vacuumed away. Cells were counted using Beckman Coulter Machine (Beckman coulter Canada Inc., Burlington, ON, Canada).
Alkaline phosphatase (ALP) activity assay
The colorimetric assay was used to determine HUCPV-Cs alkaline phosphatase (ALP) activity at the specified time points (at day 1,7 and 14 consecutively) after application of LIPUS for 10 minutes and compared with the control group. The ALP was used as the biochemical marker for osteogenic cell differentiation [37-52]. The PBS was used to wash the cells which were then lysed with 2 mL of ALP buffer per well (0.5 M 2-amino-2-methyl-1-propanol and 0.1% Triton- X-100, pH 10.5). One mL of lysed cells was used for DNA quantification assay two hours after the lysis. The ALP buffer was added in 1mg/ml to Phosphatase substrate (p-nitrophenyl phosphate) (Sigma, Oakville, ON, Canada) (1:1) ratio. 100 μl of substrate mixture and 100 μl of lysed cells were loaded to each well into 96 well plates for a final concentration of 1 mg/mL. The changes in optical density (absorbance, 405 nm) were specified in a multi-well plate reader (ELX800 Universal Microplate Reader, Bio-Tek Instruments, Inc. in Winooski, Vermont, USA.) at periodic intervals 5, 10, 15, 30 minutes.
Cell proliferation and DNA quantification assay
Measurement of DNA amount was performed using 1 mL of the lysed cell solution with the CyQUANT Cell proliferation kit (Molecular Probe, Invitrogen, Burlington, ON, Canada). Measurement of DNA quantity was performed by the CyQUANT cell proliferation kit assay (Molecular Probe, Invitrogen, Burlington, ON, Canada). Cell proliferation was determined by comparing cell’s DNA content for treated samples with untreated controls. The CyQUANT kit protocol requires binding of the cell with the dye solution, incubation for 30–60 minutes, and then measurement of fluorescence was performed in a microplate reader (Fluoroskan Ascent, Thermo Labsystems, Finland). The assay was designed to generate a linear analytical response in a 96-well microplate (Molecular Probe, Invitrogen, Burlington, ON, Canada). The DNA standard provided with the CyQUANT kit was used to determine the DNA concentrations in each cell group. Quanification of DNA used a fluorescence plate reader (excitation at 480 nm; emission at 527 nm) in accordance with the manufacturer’s instructions.
Immunophenotyping using flow-cytometry analysis
Cell surface antigen phenotyping assay was used for characterization of the HUCPVCs at passage 4 on days 1, 7 and 14 after pulsed with LIPUS and compared with control (sham) group. The following cell-surface epitopes were labeled with anti-human antibodies: CD31(PECAM-1) fluorescein isothiocyanate (FITC, BD Biosciences, Mississauga, ON, Canada), CD34-R-phycoerythrin (R-PE, BD Biosciences, Mississauga, ON, Canada), CD45-phycoerythrin (PE, BD Biosciences, Mississauga, ON, Canada), CD90 (Thy1) R-phycoerythrin (R-PE, BD Bioscience, Mississauga, ON, Canada), MHC I (HLA-A,B,C) R-phycoerythrin (RPE, BD Bioscience, Mississauga, ON, Canada), and MHC II (HLA-- DR) fluorescein isothiocyanate (FITC, BD Biosciences, Mississauga, ON, Canada) (Becton Dickinson; Beckman Coulter, Mississauga, ON, Canada), FITC-conjugated Isotype-mouse IgGa1 and PEconjugated Isotype-mouse IgGk1 served as secondary antibodies (control antibodies). A total of 10,000 labeled cells were acquired and analyzed using a FACScan flow cytometer CellQuest software (Becton Dickinson, Mississauga, ON, Canada). Details about these markers are described in Table 1. The HUCPVCs were suspended and prepared using standard direct staining protocols [40,41].
| Markers | Description |
|---|---|
| CD90 | Mesenchymal stromal cell marker |
| CD31 | Endothelial cell marker |
| CD34 | Hematopoietic cells and vascular endothelium marker |
| CD45 | Differentiated hematopoietic cell marker |
| MHC I | Recognized during graft rejection and found on all nucleated cells |
| MHCII | A marker for B-lymphocytes, macrophages and dendritic cells |
Table 1: Description of cell surface markers.
Quantitative real time-PCR analysis (Q-PCR)
Total RNA was extracted from each triplicate group of both LIPUS treated and sham (control) groups using the RNeasy Mini Kit (Qiagen, Mississauga, ON, Canada). Fluorometric quantification of RNA samples was performed at 260 nm using SYBRgreen (Molecular Probes, OR, USA), according to the manufacturer’s recommendation. One μg of the total RNA was used to synthesize single stranded DNA using the Omniscript Reverse Transcription kit (Qiagen, Mississauga, ON, Canada). The primers for real-time PCR were designed with Primer Express 2.0 software from Applied Biosystems (AB, Foster City, CA, USA.). TaqMan®Gene Expression Assays was used to perform RTPCR reactions (Applied Biosystems AB, Foster City, CA, USA.). The TaqMan®MGB probes and primers were mixed to a target concentration of 18 μM for each primer and 5 μM for the probe and the amplifications were carried out in a final reaction volume of 10 μl. Gene’s assays ID and gene’s symbols are explained in Table 2 the reaction mixtures were aliquoted into 96-well ABI reaction plate. The plates were then placed in an ABI Prism 7500 fast system V 1.4.0 Applied Bio-system q-PCR machine under the following conditions: stage 1 consisted of 95°C for 10 min; stage 2 consisted of 40 cycles of 95°C for 15 s, followed by 60°C for 1 min. The q-PCR data were analyzed with SDS 7500 Fast system V.2.01 software (AB, Foster City, CA, USA.).
| Gene Name | Gene Symbol | Assay ID |
|---|---|---|
| Endogenous Control Human GAPDH (Glyceraldehyde 3-phosphate dehydrogenase) | GAPDH | 4333764F |
| Osteocalcin (OCN) | BGLAP | Hs00609452_g1 |
| Osteopontin (OPN) | SPP1 | Hs00959009_m1 |
| Proliferating cell nuclear antigen (PCNA) | PCNA | Hs99999177_g1 |
| Nucleostemin (NST) | GNL3 | Hs00205071_m1 |
Table 2: Genes used for qPCR analysis.
Statistical analysis
Multi variate analysis of variance MANOVA was used to compare the expansion capacities of treated (LIPUS) group and control (sham) group using SPSS software package (version 16.0; SPSS Inc., Chicago, IL, USA). Analysis of the flow-cytometry data and qPCR data were completed using two-way ANOVA and the differences were statistically considered significant at (p < 0.05).
Results
The HUCPV-Cs were assessed on days 1, 7 and 14 after application of LIPUS as well as control group. The cell count in the LIPUS treated group was decreased on days 1 and 14, however, an increase noted on day 7 but not statistically significant.
There was no difference in cell proliferation assay as reflected by DNA content equalized with ALP in the LIPUS treated group (p < 0.9).
During osteogenic differentiation, no significant difference in DNA content could be detected between samples treated with LIPUS for 10 minutes per day and the untreated control group. DNA content was 0.5 fold higher on day 7 in the LIPUS treated group (0.018 ± 0.003), whereas it was lower on day 14 in the LIPUS treated group (0.015 ± 0.006) compared with the control group.
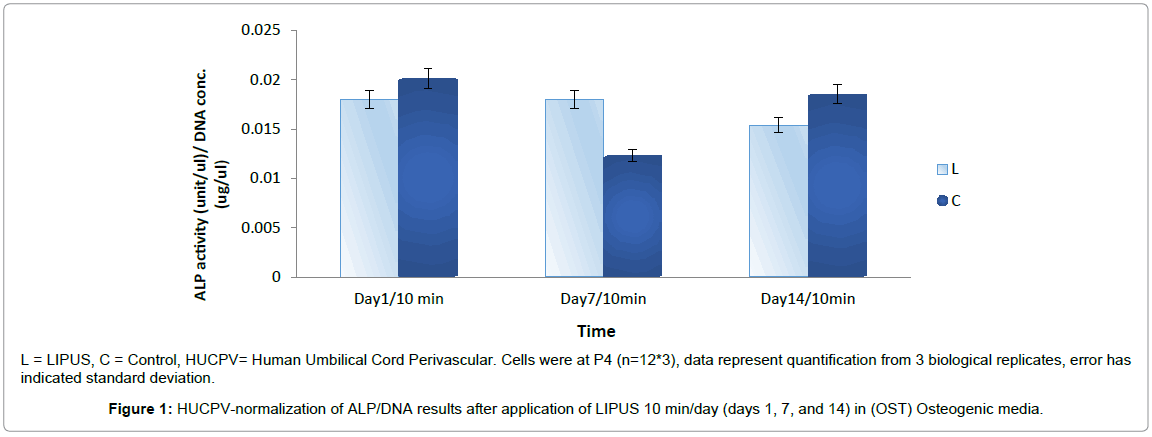
HUCPV-Cs expressed a non-significant increase of ALP activity in the LIPUS treated group compared to the control group (p < 0.9). ALP activity was slightly reduced on day 1 (0.018 ± 0.006), higher on day 7 (0.018 ± 0.003), and slightly lower on day 14 (0.015 ± 0.006) in the LIPUS treated group compared to the control group (Figure 1).
L = LIPUS, C = Control, HUCPV= Human Umbilical Cord Perivascular. Cells were at P4 (n=12*3), data represent quantification from 3 biological replicates, error has indicated standard deviation.
Figure 1: HUCPV-normalization of ALP/DNA results after application of LIPUS 10 min/day (days 1, 7, and 14) in (OST) Osteogenic media.
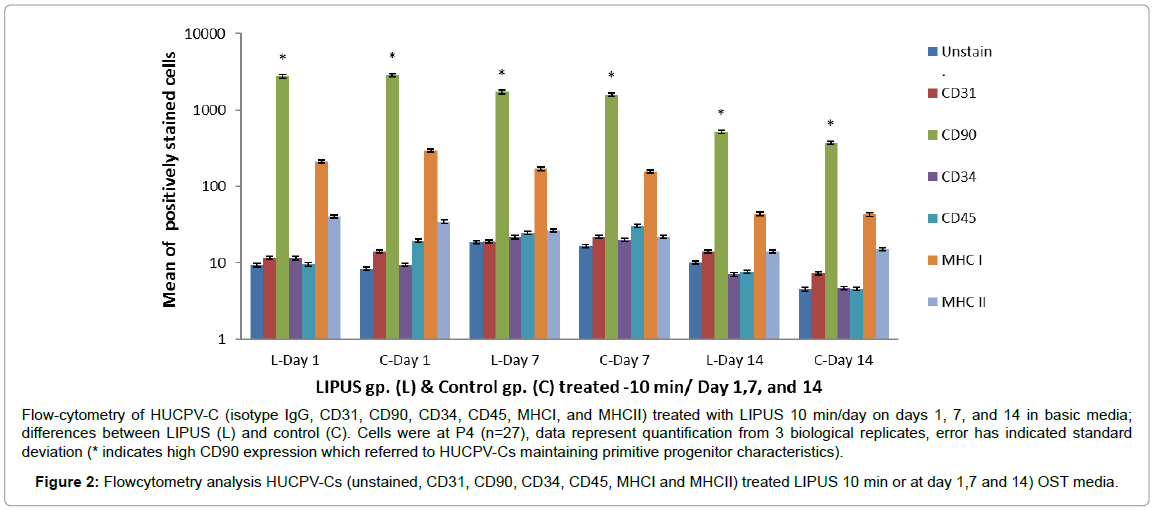
Immunophenotyping (FACS) was performed to analyze cell surface markers on HUCPV-Cs at passage 4. Cells were gated according to size and expressed surface markers. HUCPV-Cs were negative for CD31 (found on endothelial cells, platelets, macrophages) and MHCII [HLA-DR]. MHCII antigens are cell surface markers involved in graft-versus-host disease and the rejection of tissue transplants in HLA mismatched donors. HUCPV-Cs were also negative for CD34 (a hematopoietic stem cell marker) and CD45 (leukocyte common antigen). On other hand, HUCPV-Cs were strongly positive for CD90 (a mesenchymal progenitor–specific marker) and moderately positive for MHCI [HLA-A, B, C] (recognized during graft rejection, found in all nucleated cells). HUCPV-Cs in the LIPUS treated group expressed a high level of CD90 on day 14 compared with control (Figure 2 and Table 3).
Flow-cytometry of HUCPV-C (isotype IgG, CD31, CD90, CD34, CD45, MHCI, and MHCII) treated with LIPUS 10 min/day on days 1, 7, and 14 in basic media; differences between LIPUS (L) and control (C). Cells were at P4 (n=27), data represent quantification from 3 biological replicates, error has indicated standard deviation (* indicates high CD90 expression which referred to HUCPV-Cs maintaining primitive progenitor characteristics).
Figure 2: Flowcytometry analysis HUCPV-Cs (unstained, CD31, CD90, CD34, CD45, MHCI and MHCII) treated LIPUS 10 min or at day 1,7 and 14) OST media.
| Markers / OST | Day 1 | Day 7 | Day 14 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| L Mean ± SD | C Mean ±SD | p value | L Mean ± SD | C Mean ± SD | p value | L Mean ±SD | C Mean ± SD | pvalue | |
| IsotypeIgG | 9.3±2.9 | 8.4±2.5 | 0.5 | 18.5±4.6 | 16.7±9.3 | 0.6 | 10.1±9.6 | 4.5±2.5 | 0.6 |
| CD31 | 11.6±3.4 | 14±1.8 | 0.3 | 19±2.5 | 22±14.4 | 0.3 | 14.1±8.2 | 7.3±2.2 | 0.3 |
| CD90 | 2766.7±156.9 | 2854.5±549.4 | 0.7 | 1731.3±732.9 | 1601.1±771.5 | 0.9 | 516.7±292.5 | 370.7±162.5 | 0.9 |
| CD34 | 11.5±6.2 | 9.4±1.5 | 0.9 | 21.6±10.9 | 19.8±12.4 | 0.9 | 7±4.9 | 4.6±1.6 | 0.9 |
| CD45 | 9.5±3.6 | 19.4±15.1 | 0.3 | 24.4±14.6 | 30.5±30 | 0.9 | 7.6±5.4 | 4.5±1.1 | 0.9 |
| MHC I | 211.5±66.5 | 294.5±177.6 | 0.7 | 170.3±164.7 | 155.9±157.7 | 0.9 | 43.6±24.8 | 42.7±22.7 | 0.9 |
| MHC II | 40.1±25.5 | 34.4±27.2 | 0.7 | 26.5±3.4 | 21.9±16.2 | 0.8 | 13.9±7 | 14.9±8.7 | 0.8 |
Table 3: Comparison of cell surface markers expression of flow-cytometry results of HUCPV-SC (isotype IgG, CD31, CD90, CD34, CD45, MHCI, and MHCII) treated with LIPUS 10 min/day on days 1, 7, and 14: difference between LIPUS (L) and control (C) in Osteogenic media. The results expressed as Mean ± SD.
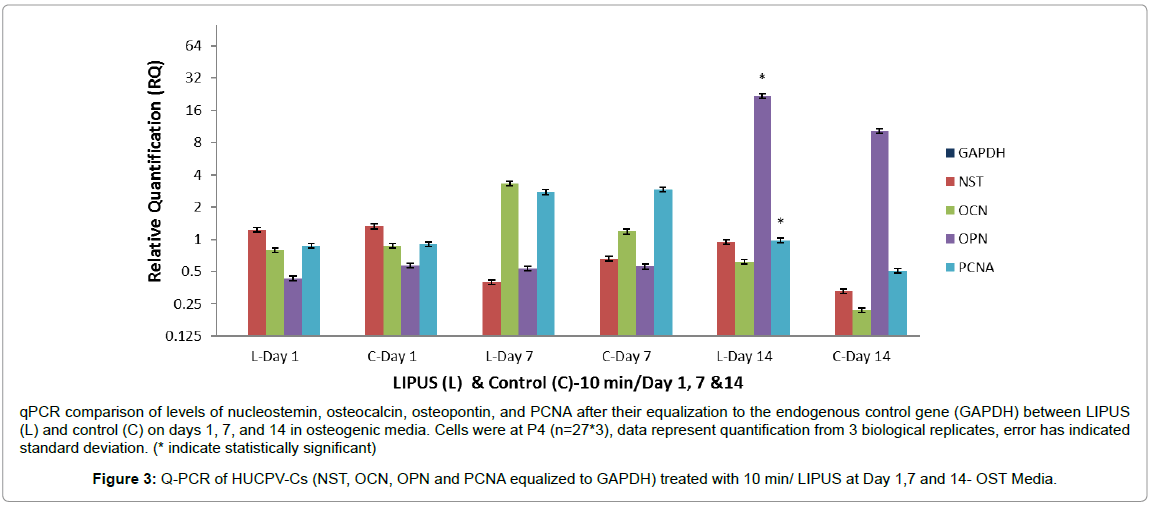
We further investigated our original hypothesis, that LIPUSexpanded HUCPV-Cs will maintain their osteogenic differentiation potential, by assessing the expression of nucleostemin, PCNA, OCN, and OPN after equalization to the endogenous control gene GAPDH. Nucleostemin is a marker of undifferentiated human mesenchymal stromal stem cells and is involved in regulation of MSC proliferation [42]. HUCPV-Cs expressed lower levels of nucleostemin in the LIPUS treated group on days 1 and 7 compared to the control, with a nonsignificant higher expression on day 14 (Table 4). On the other hand, the level of PCNA was significantly higher in the LIPUS treated group on day 14 (p < 0.001).
| Genes / OST | Day 1 | Day 7 | Day 14 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| L Mean ±SD | C Mean ±SD | p value | L Mean ±SD | C Mean ±SD | p Value | LMean ±SD | CMean ±SD | p value | |
| GAPDH | 0.00 ±0.00 | 0.00 ±0.00 | 0.8 | 0.00 ±0.00 | 0.00 ±0.00 | 0.9 | 0.00 ±0.00 | 0.00±0.00 | 0.9 |
| NST | 1.23 ±0.16 | 1.32 ±0.19 | 0.6 | 0.39 ±0.42 | 0.66 ±0.57 | 0.9 | 0.95 ±0.10 | 0.33 ±0.05 | 0.9 |
| OCN | 0.79 ±0.19 | 0.87 ±0.12 | 0.7 | 3.33 ±3.54 | 1.18 ±1.39 | 0.08 | 0.62 ±0.29 | 0.22 ±0.07 | 0.9 |
| OPN | 0.43 ±0.21 | 0.57 ±0.42 | 0.4 | 0.54 ±0.40 | 0.56 ±0.65 | 0.9 | 21.84 ±15.64 | 10.25 ±3.61 | 0.001 |
| PCNA | 0.87 ±0.15 | 0.91 ±0.04 | 0.4 | 2.76 ±2.01 | 2.92 ±3.62 | 0.9 | 0.98 ±0.10 | 0.51 ±0.05 | 0.001 |
Table 4: Comparison of Q-PCR expression of cell genes nucleostemin, osteocalcin, osteopontin, and PCNA after their equalization to the endogenous control gene (GAPDH) treated with LIPUS (L) 10 min/ day on days 1, 7, and 14: difference between LIPUS (L) and Control (C) in Osteogenic Media (OST). The results expressed as Mean ±SD.
The levels of OCN expression were approximately 0.2 fold lower in the LIPUS treated group on day 1, 1.5 fold higher on day 7, and 0.5 fold higher on day 14. These responses were, however, statistically nonsignificant. The level of OPN was 1 fold higher on day 14 (p < 0.001), whereas it was 0.2 fold lower on day 1 and almost comparable to the control group on day 7. These findings suggest that LIPUS treatment for 10 min/day may enhance osteogenic differentiation of HUCPV-Cs on day 14 and beyond (Figure 3 and Table 4).
qPCR comparison of levels of nucleostemin, osteocalcin, osteopontin, and PCNA after their equalization to the endogenous control gene (GAPDH) between LIPUS (L) and control (C) on days 1, 7, and 14 in osteogenic media. Cells were at P4 (n=27*3), data represent quantification from 3 biological replicates, error has indicated standard deviation. (* indicate statistically significant)
Figure 3: Q-PCR of HUCPV-Cs (NST, OCN, OPN and PCNA equalized to GAPDH) treated with 10 min/ LIPUS at Day 1,7 and 14- OST Media.
Discussion
MSCs have shown to be differentiated into osteoblastic lineage [16,18]. Osteogenic differentiation of MSCs was established in culture media containing ascorbic acid, β-glycerophosphate, and dexamethasone. It has been demonstrated previously that HUCPV-Cs are capable to be differentiated into osteogenic lineage in vitro after incubation in osteogenic media for 5, 21, and 28 days [16,18]. The stimulatory effect of LIPUS has been documented in many studies using a variety of cell lineages such as osteoblasts, chondrocytes, and marrow-derived stromal cells [43-46].
Our results showed a non-significant increase in HUCPV-Cs osteogenic differentiation capacity after 1 day of LIPUS application. On day 7 of LIPUS application, there have been increase in some osteogenic markers, namely OCN and ALP, and there was a significant increase in OPN on day 14. No significant differences in cell count, DNA content, or immunophenotypic characteristics were detected between the LIPUS treated preparation and a sham treated control.
DNA content, ALP activity, and calcium content were used as alternate measures for cellular activities in some experimental studies. The expression of these has been shown to decrease by mechanical stress, such as stretching and loading [47]. Similarly, it has been shown that intermittent loading of mechanical stress reduces the activation of mechanosensitive cation channels on osteoblast-like cells [48]. Intermittent cyclic loading has been used as a form of applied mechanical stress [47,48]. Some of these findings were consistent with our results; that is, the down-regulation of some cellular markers after exposure to LIPUS [47-50].
Non-significant increases of CD90 and nucleostemin on day 14 were noted in the LIPUS treated group. In addition, non-significant changes in levels of OCN were observed: OCN was approximately 0.2 fold lower on day 1, 1.5 fold higher on day 7, and 0.5 fold higher on day 14. Statistically significant higher expression of PCNA and OPN in the LIPUS treated group compared to the control was observed on day 14. These findings suggest that the stimulatory effect of LIPUS application to upregulate OPN gene expression in HUCPV-Cs occurs after 14 days of daily. Lee et al. reported that LIPUS enhances cell viability by increasing expression of cell viability related genes such as PCNA [51-53]. Biomechanical stimulation and LIPUS were effective tools in improving repair of damaged cells and enhance the synthesis of matrix protein [53]. It has been reported that application of LIPUS for 20 days to human mandibular fracture haematoma-derived cells (MHCs) significantly increase osteogenic gene expression and osteogenic protein [54]. Also, it has been reported that LIPUS enhances bone repair in animals, upregulate osteogenic genes expression, and significantly increase alkaline phosphatase and bone morphogenic protein after 25 days of application [55]. Furthermore, it has been shown that LIPUS stimulation has profoundly enhanced the multifunctional effect that is relevant to alveolar bone regeneration which plays an important factor in periodontal healing [56]. They reported that human alveolar bonederived mesenchymal stem cells (hABMSCs) treated with 50 mW/cm2 show a significant increase in osteogenic genes expression, alkaline phosphatase, and calcium deposit compared to untreated group after 10 minutes /day application for 3 weeks [56]. In summary, future studies may aim to investigate the possible stimulatory effect of increasing LIPUS application time to 20 minutes/day for 20 days or more for the possibility that more significant osteogenic gene expression might occur.
Conclusion
This study explored the effect of daily application of LIPUS for 10 minutes/day for 1, 7 and 14 days on the osteogenic differentiation of HUCPV-Cs. The results of this study are as follow: HUCPV-Cs treated with LIPUS demonstrate increase in CD90 level at day 14 compared to untreated group. Moreover, LIPUS application to stimulated HUCPVCs showed statistically significant increase in PCNA, and OPN gene expression on day 14, respectively. These results suggest that LIPUS has stimulatory effect on osteogenic differentiation of HUCPV-Cs especially after 14 days of application. More studies may be conducted to investigate the possible effect of different LIPUS frequencies, power or treatment time on the osteogenic differentiation of HUCPV-Cs.
Acknowledgement
This research was supported by the Fund for Dentistry, Department of Dentistry, University of Alberta, Edmonton, Canada and the Saudi government.
References
- Reyes M, Lund T, Lenvik T, Aguiar D, Koodie L, et al. (2001) Purification and ex vivo expansion of postnatal human marrow mesodermal progenitor cells. Blood 98: 2615-2625. [PubMed]
- Lee OK, Kuo TK, Chen WM, Lee KD, Hsieh SL, et al.(2004) Isolation of multipotentmesenchymal stem cells from umbilical cord blood. Blood 103:1669-1675. [PubMed]
- Anker PS, Noort WA, Kruisselbrink AB, Scherjon SA, Beekhuizen W, et al. (2003) Nonexpanded primary lung and bone marrow-derived mesenchymal cells promote the engraftment of umbilical cord blood-derived CD34(+) cells in NOD/SCID mice. Exp. Hematol. 31:881-889. [PubMed]
- Broxmeyer HE, Douglas GW, Hangoc G, Cooper S, Bard J, et al. (1989) Human umbilical cord blood as a potential source of transplantable hematopoietic stem/progenitor cells. ProcNatlAcadSci U S A 86:3828-3832. [PubMed]
- Gluckman E, Broxmeyer HA, Auerbach AD, Friedman HS, Douglas GW, et al. (1989) Hematopoietic reconstitution in a patient with Fanconi's anemia by means of umbilical-cord blood from an HLA-identical sibling. N Engl J Med321: 1174-1178. [PubMed]
- Wagner JE, Rosenthal J,Sweetman R, Shu XO, Davies SM, et al. (1996) Successful transplantation of HLA-matched and HLA-mismatched umbilical cord blood from unrelated donors: analysis of engraftment and acute graft-versus-host disease. Blood 88:795-802. [PubMed]
- Jin HJ, Park SK, Oh W, Yang YS, Kim SW, et al.(2009) Down-regulation of CD105 is associated with multi-lineage differentiation in human umbilical cord blood-derived mesenchymal stem cells. BiochemBiophys Res Commun381:676-681. [PubMed]
- Wagner W, Wein F, Seckinger A, Frankhauser M, Wirkner U, et al. (2005) Comparative characteristics of mesenchymal stem cells from human bone marrow, adipose tissue, and umbilical cord blood. ExpHematol 33:1402-1416. [PubMed]
- Kern S, Eichler H, Stoeve J, Kluter H, Bieback K(2006) Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood, or adipose tissue. Stem Cells 24:1294-1301. [PubMed]
- Erices A, Conget P, Minguell JJ (2000) Mesenchymal progenitor cells in human umbilical cord blood. Br J Haematol 109:235-242. [PubMed]
- Lee MW, Choi J, Yang MS, Moon YJ, Park JS, et al. (2004) Mesenchymal stem cells from cryopreserved human umbilical cord blood. BiochemBiophys ResCommun 320:273-278. [PubMed]
- Kestendjieva S, Kyurkchiev D, Tsvetkova G, Mehandjiev T, Dimitrov A, et al. (2008) Characterization of mesenchymal stem cells isolated from the human umbilical cord. Cell BiolInt 32:724-732. [PubMed]
- Baksh D, Yao R, Tuan RS(2007) Comparison of proliferative and multilineage differentiation potential of human mesenchymal stem cells derived from umbilical cord and bone marrow. Stem Cells 25:1384-1392. [PubMed]
- Qiao C, Xu W, Zhu W, Hu J, Qian H, et al. (2008) Human mesenchymal stem cells isolated from the umbilical cord. Cell BiolInt 32:8-15. [PubMed]
- Gang EJ, Hong SH, Jeong JA, Hwang SH, Kim SW, et al. (2004) In vitro mesengenic potential of human umbilical cord blood-derived mesenchymal stem cells. BiochemBiophys Res Commun 321:102-108. [PubMed]
- Wang HS, Hung SC, Peng ST, Huang CC, Wei HM, et al. (2004) Mesenchymal stem cells in the Wharton's jelly of the human umbilical cord. Stem Cells 22:1330-1337. [PubMed]
- Weiss ML, Medicetty S, Bledsoe AR, Rachakatla RS, Choi M, et al. Human umbilical cord matrix stem cells: preliminary characterization and effect of transplantation in a rodent model of Parkinson's disease. Stem Cells 24:781-792. [PubMed]
- Sarugaser R, Lickorish D, Baksh D, Hosseini MM, Davies JE(2005) Human umbilical cord perivascular (HUCPV) cells: a source of mesenchymal progenitors. Stem Cells 23:220-229. [PubMed]
- Gluckman E, Rocha V, Boyer-Chammard A, Locatelli F, Arcese W, et al. (1997) Outcome of cord-blood transplantation from related and unrelated donors. Eurocord Transplant Group and the European Blood and Marrow Transplantation Group. NEngl J Med 337:373-381. [PubMed]
- Han IS, Ra JS, Kim MW, Lee EA, Jun HY, et al. (2003) Differentiation of CD34+ cells from human cord blood and murine bone marrow is suppressed by C6 beta-chemokines. Mol Cells 15:176-180. [PubMed]
- Kim JW, Kim SY, Park SY, Kim YM, Kim JM, et al. Mesenchymal progenitor cells in the human umbilical cord. Ann Hematol 83:733-738. [PubMed]
- Goodwin HS, Bicknese AR, Chien SN, Bogucki BD, Quinn CO, et al. (2001)Multilineage differentiation activity by cells isolated from umbilical cord blood: expression of bone, fat, and neural markers. Biol Blood Marrow Transplant 7:581-588. [PubMed]
- Heckman JD, Ryaby JP, McCabe J, Frey JJ, Kilcoyne RF(1994) Acceleration of tibial fracture-healing by non-invasive, low-intensity pulsed ultrasound. J Bone Joint Surg Am 76:26-34. [PubMed]
- Kristiansen TK, Ryaby JP, McCabe J, Frey JJ, Roe LR (1997) Accelerated healing of distal radial fractures with the use of specific, low-intensity ultrasound. A multicenter, prospective, randomized, double-blind, placebo-controlled study. J Bone Joint Surg Am 79:961-973. [PubMed]
- Mayr E, Rudzki MM, Rudzki M, Borchardt B, Hausser H, et al.(2000) Does low intensity, pulsed ultrasound speed healing of scaphoid fractures? HandchirMikrochirPlastChir32:115-122. [PubMed]
- Nolte PA, van der KransA, Patka P, Janssen IM, Ryaby JP, et al. (2001) Low-intensity pulsed ultrasound in the treatment of nonunions. J Trauma 51:693-702. [PubMed]
- Leung KS, Lee WS, Cheung WH, Qin L(2004) Lack of efficacy of low-intensity pulsed ultrasound on prevention of postmenopausal bone loss evaluated at the distal radius in older Chinese women. ClinOrthopRelat Res 427:234-240. [PubMed]
- Tsumaki N, Kakiuchi M, Sasaki J, Ochi T, Yoshikawa H(2004) Low-intensity pulsed ultrasound accelerates maturation of callus in patients treated with opening-wedge high tibial osteotomy by hemicallotasis. J Bone Joint Surg Am 86-A:2399-2405. [PubMed]
- Gebauer D, Mayr E, Orthner E, Ryaby JP(2005) Low-intensity pulsed ultrasound: effects onnonunions. Ultrasound Med Biol31:1391-1402. [PubMed]
- Gold SM, Wasserman R (2005) Preliminary results of tibial bone transports with pulsed low intensity ultrasound (Exogen). JOrthop Trauma 19:10-16. [PubMed]
- Ricardo M(2006) The effect of ultrasound on the healing of muscle-pediculated bone graft in scaphoid non-union. IntOrthop 30:123-127. [PubMed]
- Schmelz A, Friedrich A, Kinzl L, Einsiedel T (2006) Low intensity pulsed ultrasound (LIPUS) is shortening healing time following callus distraction for bony defects of the tibia. AktuelleTraumatol36:149-158.
- Nomura S, Takano-Yamamoto T (2000) Molecular events caused by mechanical stress in bone. Matrix Biol 19:91-96. [PubMed]
- Bandow K, Nishikawa Y, Ohnishi T, Kakimoto K, Soejima K, et al. (2007) Low-intensity pulsed ultrasound (LIPUS) induces RANKL, MCP-1, and MIP-1beta expression in osteoblasts through the angiotensin II type 1 receptor. J Cell Physiol211:392-398. [PubMed]
- Ikai H, Tamura T, Watanabe T, Itou M, Sugaya A, et al. (2008) Low-intensity pulsed ultrasound accelerates periodontal wound healing after flap surgery. J Periodontal Res 43:212-216. [PubMed]
- Eberson CP, Hogan KA, Moore DC, Ehrlich MG (2003) Effect of low-intensity ultrasound stimulation on consolidation of the regenerate zone in a rat model of distraction osteogenesis. J PediatrOrthop 23:46-51. [PubMed]
- Leung KS, Cheung WH, Zhang C, Lee KM, Lo HK(2004) Low intensity pulsed ultrasound stimulates osteogenic activity of human periosteal cells. ClinOrthop 418:253-259. [PubMed]
- Martini L, Giavaresi G, Fini M, Torricelli P, de Pretto M, et al. (2003) Effect of extracorporeal shock wave therapy on osteoblastlike cells.ClinOrthopRelat Res 413:269-280. [PubMed]
- Owen TA, Aronow M, Shalhoub V, Barone LM, Wilming L, et al. Progressive development of the rat osteoblast phenotype in vitro: reciprocal relationships in expression of genes associated with osteoblast proliferation and differentiation during formation of the bone extracellular matrix. J CellPhysiol 143:420-430. [PubMed]
- Holmes K, Lantz LM, Fowlkes BJ, Schmid I, Giorgi JV(2001) Preparation of cells and reagents for flow cytometry. CurrProtocImmunol. [PubMed]
- Malley A, Stewart CC, Stewart SJ, Waldbeser L, Bradley LM, et al. (1988) Flow cytometric analysis of I-J expression on murine bone marrow-derived macrophages. J LeukocBiol43:557-565. [PubMed]
- Kafienah W, Mistry S, Williams C, Hollander AP (2006) Nucleostemin is a marker of proliferating stromal stem cells in adult human bone marrow. Stem Cells 24:1113-1120. [PubMed]
- Sun JS, Hong RC, Chang WH, Chen LT, Lin FH, et al. (2001) In vitro effects of low-intensity ultrasound stimulation on the bone cells. J Biomed Mater Res57:449-456. [PubMed]
- Parvizi J, Wu CC, Lewallen DG, Greenleaf JF, Bolander ME (1999) Low-intensity ultrasound stimulates proteoglycan synthesis in rat chondrocytes by increasing aggrecan gene expression. J Orthop Res17:488-494. [PubMed]
- Zhang ZJ, Huckle J, Francomano CA, Spencer RG (2003) The effects of pulsed low-intensity ultrasound on chondrocyte viability, proliferation, gene expression, and matrix production. Ultrasound Med Biol 29:1645-1651. [PubMed]
- Naruse K, Mikuni-Takagaki Y, Azuma Y, Ito M, Oota T, et al. (2000) Anabolic response of mouse bone-marrow-derived stromal cell clone ST2 cells to low-intensity pulsed ultrasound. BiochemBiophys Res Commun 268:216-220. [PubMed]
- Winter LC, Walboomers XF, Bumgardner JD, Jansen JA(2003) Intermittent versus continuous stretching effects on osteoblast-like cells in vitro. J Biomed Mater Res A 67:1269-1275. [PubMed]
- Duncan RL,Hruska KA(1994) Chronic, intermittent loading alters mechanosensitive channel characteristics in osteoblast-like cells. Am JPhysiol267:F909-16. [PubMed]
- Karahuseyinoglu S, Cinar O, Kilic E, Kara F, Akay GG, et al. (2007) Biology of stem cells in human umbilical cord stroma: in situ and in vitro surveys. Stem Cells 25:319-331. [PubMed]
- Yoon JH, Roh EY, Shin S, Jung NH, Song EY, et al.(2009) Introducing pulsed low-intensity ultrasound to culturing human umbilical cord-derived mesenchymal stem cells. BiotechnolLett31:329-335. [PubMed]
- Marvel S, Okrasinski S, Bernacki SH, Loboa E, Dayton PA (2010) The development and validation of a LIPUS system with preliminary observations of ultrasonic effects on human adult stem cells. IEEE Trans UltrasonFerroelectrFreq Control57:1977-1984. [PubMed]
- Mostafa NZ, Uludag H, Dederich DN, Doschak MR, El-Bialy TH(2009) Anabolic Effects of Low Intensity Pulsed Ultrasound on Gingival Fibroblasts.Arch Oral Biol54: 743-748. [PubMed]
- Lee HJ, Choi BH, Min BH, Park SR (2007) Low-intensity ultrasound inhibits apoptosis and enhances viability of human mesenchymal stem cells in three-dimensional alginate culture during chondrogenic differentiation. Tissue Eng13:1049-1057. [PubMed]
- Huang W, Hasegawa T, Imai Y, Takeda D, Akashi M (2015) Low-intensity pulsed ultrasound enhances bone morphogenetic protein expression of human mandibular fracture haematoma-derived cells. Int J Oral MaxillofacSurg44:929-935. [PubMed]
- Fávaro-Pípi E, Bossini P, de Oliveira P, Ribeiro JU, Tim C, et al. (2010) Low-Intensity Pulsed Ultrasound Produced an Increase of Osteogenic Genes Expression During the Process of Bone Healing in Rats. Ultrasound Med Biol36:2057-2064. [PubMed]
- Lim K, Kim J, Seonwoo H, Park S, Choung P, et al. (2013) In vitro effects of low-intensity pulsed ultrasound stimulation on the osteogenic differentiation of human alveolar bone-derived mesenchymal stem cells for tooth tissue engineering. Biomed ResInt 2013:269724. [PubMed]
Copyright: © 2016 Aldosary TA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.