Indexed In
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- Ulrich's Periodicals Directory
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Proquest Summons
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Review Article - (2023) Volume 14, Issue 3
Next Generation Sequencing of Human Red Cell Antigens Kell, Kidd, and Duffy for Routine Clinical Investigations and Donor Screening: A Systematic Review and Meta-Analysis
Hua HC and Jackson DE*Received: 01-Feb-2023, Manuscript No. JBDT-23-19746; Editor assigned: 03-Feb-2023, Pre QC No. JBDT-23-19746 (PQ); Reviewed: 23-Feb-2023, QC No. JBDT-23-19746; Revised: 03-Mar-2023, Manuscript No. JBDT-23-19746 (R); Published: 10-Mar-2023, DOI: 10.4172/2155-9864.23.14.552
Abstract
The emergence of Next-Generation Sequencing (NGS) allows replacement over current molecular testing for red cell antigens in routine practice of transfusion laboratories in determining individual phenotypes. The high throughput platforms enhance the likelihood of donor matching by extended blood group genotyping and meanwhile explore novel and rare polymorphisms, potentially assisting in large-pooled donor screening and well-typed inventory build- up in blood banks. To reconcile NGS application in this field, this systematic review and meta-analysis of the literature was conducted to examine whether NGS has adequate grounds to replace current SNV-based genotyping. Overall, 362 samples in 6 eligible studies were studied upon screening through inclusion/exclusion criteria. Concordance analyses between NGS platforms and serology or other molecular typing methods on Kell, Kidd, and Duffy genes were performed to investigate the accuracy of NGS in donor phenotype prediction. The pooled proportion agreement for the 6 included studies on the overall concordance between NGS and comparators were 0.987 (95% CI, 0.975 to 0.996; P<0.001) for Kell, 0.984 (95% CI, 0.968 to 0.994; P<0.001) for Kidd, and 0.986 (95% CI, 0.973 to 0.995; P<0.001) for Duffy genotyping. Our results demonstrated accurate typing of Kell, Kidd, and Duffy genes in blood samples by NGS in conjunction with its ability in the unprecedented evaluation of novel and complex structural variants, though technological and methodological hurdles still exist. As such, NGS is still a complementary tool to serology with its potential manifested by further studies and advances in sequencing platforms.
Keywords
Kell; Kidd; Duffy; NGS- Next Generation Sequencing; Genotyping; Blood donor; Red cell antigen
Introduction
Red blood cell antigen
The membrane of every erythrocyte is coated with distinct antigens, glycoproteins, glycolipids, or proteins [1]. These antigens are integrally linked to different components for various cell functions such as membrane transporters, chemokine receptors, and cell adhesion molecules [2]. The combination of antigens expressed on red cells defines an individual's blood group. To date, the International Society of Blood Transfusion (ISBT) working party for Red Cell Immunogenetics and blood group terminology allocated more than 300 Red Blood Cell antigens (RBC) genetically revealed by 48 genes to 43 blood group systems [1]. Under this terminology, one blood group system corresponds to antigens determined by one single gene or those with closely related loci and thus is genetically different [3]. Among all discovered blood group systems, 22 can provoke severe transfusion reactions, including the most prevalent ABO, Rhesus, Kell, Kidd, Duffy, and MNS blood groups.
Kell, kidd, and duffy blood group systems
The Kell gene (KEL), with its locus on chromosome 7q33, encodes an endothelin-3-converting transmembrane enzymatic glycoprotein. The Kell system has been discovered with 31 antigens due to the high chromosomal polymorphism [4].
The Kidd antigens, Jka and Jkb, are expressed by two codominant alleles JK*A and JK*B located on chromosome 18q11-q12. The glycoproteins are responsible for transporting urea on red cells. The absence of any Kidd antigens corresponds to the rare Jk-null phenotype [5].
The Duffy system is harboured by two exons on chromosome 1q22-q23 genomic region. The two major alleles, FY*A and FY*B, of the Duffy gene (Fy) encode the Fya and Feb antigens, multipass transmembrane glycoproteins functioning as receptors for cytokine secreted during inflammation [5].
Prevention of alloimmunisation against red blood cell antigens
Red cell antigens are of varying clinical significance in transfusion and obstetric medicine with their immunogenicity, in which the introduction of foreign antigens from donor material or fetal blood evokes the production of corresponding antibodies and the occurrence of sensitizing events leading to alloimmunisation [6]. Alloantibodies developed may lead to the destruction of red cells and cause complications of varying severity of Hemolytic Transfusion Reactions (HTR) or Hemolytic Disease of the Fetus and Newborn (HDFN) [7]. Studies reported the risks of allogenic blood transfusion with the prevalence of alloantibodies in patients as a result of red cell transfusion being up to 50% [6]. A Canadian study revealed the estimated risk of acute HTR to be 1:13 000 and delayed HTR to be 1:9 000 as associated with transfusion of cellular blood components [8]. Thus, giving out prophylactically matched donor’s blood to patients is essential to minimize the risks of developing atypical antibodies following clinical problems.
Clinical practices in red blood cell antigen typing
Red blood cell transfusion is a typical clinical procedure upon the indication of anemia or impaired oxygen delivery. Complex isoimmunisation imposes great difficulty in finding compatible blood for patients and thus remains a complication in the blood bank service. Serology test is a conventional method in blood typing for pretransfusion purposes to detect any hemagglutination resulting from antigen-antibody reactions [9]. Nevertheless, phenotyping exerts certain limitations including immunoglobulin from treatment coating on red cells, transfusion history, discrepancies, and limited availability of antisera [10]. Thus, molecular techniques targeting polymorphisms and mutations in the prediction of specific red blood cell antigen expression and corresponding blood groups become a more dependable tool in conjunction with the traditional serological typing method in recent years [11].
A Single Nucleotide Polymorphism (SNP) is the most prevalent variation that occurs with an alteration in amino acid in the peptide sequence leading to different antigen expressions [12]. Several molecular methodologies for erythrocyte genotyping have been introduced based on Polymerase Chain Reaction (PCR) of genetic sequences. Determination of SNPs of erythrocyte antigens can be achieved by a variety of molecular techniques such as the PCRSequence Specific Primer (PCR-SSP) using a specific primer to flank and detect nucleotide sequence of the polymorphic alleles and the PCR-Restriction Fragment Length Polymorphism (PCR-RFLP) is using restriction enzymes [13].
Next generation sequencing
Next-Generation Sequencing (NGS) usually refers to the second generation in sequencing techniques evolving from the Sanger-based method. NGS offers advantages beyond classic techniques including cost-effectiveness, high throughput capacity for massively parallel sequencing, and relatively short reads [6]. There are currently various commercialized NGS platforms such as ThermoFisher Scientific, Illumina, Roche Life Sciences, and Applied Biosystems [14]. NGS can be applied in various levels of sequencing, from a selected region of gene interest to Whole Genome Sequencing (WGS) and Whole Exome Sequencing (WES). The capacity allows blood group typing and novel variants discovery responsible for different blood group phenotypes [12]. The advances enable extended profiling of blood samples for massive donor screening and cross-matching in transfusion.
Blood group determination by laboratory analyses has a prerequisite role in routine transfusion to eliminate the risk of clinical complications due to alloimmunisation. With the potential of using the extraordinarily high-throughput NGS technology in blood group determination and massive donor screening, it is important to consider the accuracy and thus reliability of this newly emerged technique before putting it as a supplemental approach to the routinely used serological typing and genotyping.
The most important blood group systems, ABO, and Rhesus are usually emphasized in studies. Nonetheless, the investigation on the next clinically significant blood groups, Kell, Kidd, and Duffy across the topic of NGS platform accuracy and discrepancy is limited whilst hemagglutination tests can be time-consuming and hard to interpret in patients especially those who have recently transfused with blood [15, 16]. Under these circumstances, the study aimed to perform a systematic review and meta-analysis of studies to examine the accuracy of current NGS platforms as compared to the results from serological phenotyping or classical genotyping particularly in Kell, Kidd, and Duffy blood group systems by comparing the concordance between methods, and thus to evaluate the feasibility of adopting NGS as a supplemental tool in donor blood group typing in the future to ease the burden of transfusion service.
Materials and Methods
Study design
This systematic review was conducted and presented upon adherence to the guidelines from the Preferred Reporting Item for Systematic Review and Meta-Analysis (PRISMA) [17].
Selection criteria
Types of studies: An extensive review of all prospective and retrospective observational studies was conducted. All fully accessible publications in English were considered potentially eligible, irrespective of publication date. Case studies were excluded from analysis due to inadequate sample size. Aside from original research, conference abstracts, book chapters, letters, inaccessible articles, and systematic reviews, were also excluded.
Inclusion and exclusion criteria
Studies with participants of different ethnicities were considered eligible for inclusion. Studies that analyzed samples from blood donors, reference laboratories, and genome project databases were also eligible for analysis.
The predefined criteria for eligible studies for inclusion were:
• The use of molecular genotyping of Kell, Kidd, and Duffy red cell antigens using a next-generation DNA sequencing platform, regardless of the exact NGS platform used.
• The comparison of red cell antigen genotyping results of samples to other typing methods, such as serology, PCR-SSP, and Sanger sequencing. Articles that investigate genotyping results of other blood group systems were not excluded considering that targeted blood group systems were also evaluated, and findings could be extracted correspondingly. However, articles with emphasis on only the unrelated blood group antigens or other human antigens were excluded. Review articles or articles that merely focus on disease diagnosis or novel variants detection were not considered eligible.
Outcome measurement
Studies that illustrated donor genotyping results generated from NGS and comparator arms in parallel were explored and counted as eligible for inclusion in this review. Studies without reporting the number of concordant events in findings were excluded.
Search process for studies identification
According to the defined study requirements, a comprehensive search was conducted on four electronic databases: PubMed, EMBASE, Scopus, and Google Scholar (August 2012-August 2022). The retrieval terms used for the search included “human red cell antigen”, “red cell antigen”, “blood group”, “genotyping”, “NGS” and “whole genome sequencing” amongst others. A manual search of selected references for additional relevant articles was also performed.
Data retrieval and management
The author performed screening on all search results by evaluating the titles and abstracts of the studies. Full-text articles were extracted upon potential eligibility. The general characteristics (year of publication, author, geographic location, study design, NGS platform, and comparative typing methods used), population characteristics, and concordance outcomes were extracted.
Assessment of risk of bias
The possible risk of bias and the quality of the included articles was examined according to a checklist adopted from the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) to ensure high-quality presentation with and across studies [18].
Statistical analysis
For each included study, the concordance in percentage agreement between Kell, Kidd, and Duffy genotypes resulting from next-generation sequencing techniques and its serological typing or Single Nucleotide Variant (SNV)-based genotyping comparator was regarded the primary outcome of the meta-analysis. Arcsine transformation was applied on concordant proportions to stabilize the variance. Proportions were pooled using random effects, Maximum Likelihood (ML) model for weights calculation. Results were illustrated in a forest plot generated by OpenMeta [Analyst], with a respective α=0.05 significance level [19]. I2 statistics were computed for indication of the percentage of overall variation attributable to the heterogeneity across studies [20]. 95% Confidence Interval (CI) was reported and ≤ 0.05 in p-value was considered indicative of statistical significance.
Results
Selection of studies
The initial database search identified 1345 articles (PubMed, n=230; EMBASE, n=202; Scopus, n=716; Google Scholar, n=197) and 1 article from manual literation search with potential eligibility for inclusion. Of the 178 articles remained after elimination of 1168 duplicates, 993 records were excluded based on irrelevant focuses on disease diagnosis or treatment (n=388), novel allele detection (n=226), or inaccessibility (n=130) in major. Among the 175 articles assessed for eligibility, 169 records that failed to meet the criteria of using NGS techniques (n=96) and identification of Kell, Kidd, or Duffy blood groups (n=73) were excluded. Following a qualitative full-text review, 6 cohort studies concentrating on next-generation sequencing for red cell antigens genotyping including Kell, Kidd, and Duffy with a comparison of alternative typing methods were ultimately included in the systematic review and meta-analysis (Figure 1). Subject samples did not overlap among study groups upon analysis.
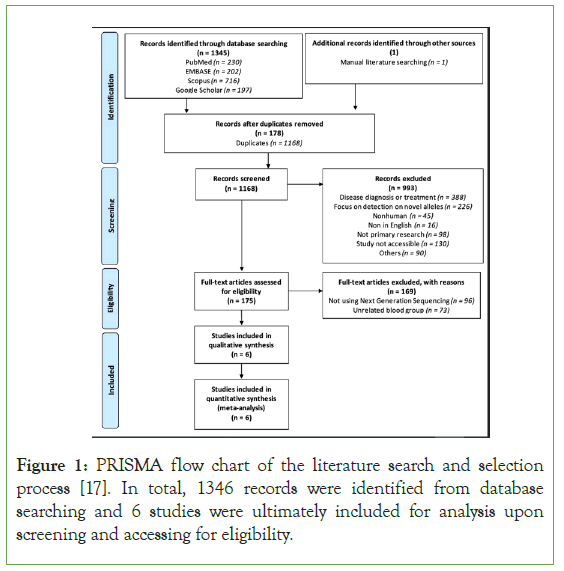
Figure 1: PRISMA flow chart of the literature search and selection process [17]. In total, 1346 records were identified from database searching and 6 studies were ultimately included for analysis upon screening and accessing for eligibility.
Quality and risk of bias assessment
Data quality of the included studies was investigated according to items from the STROBE statement as shown in Table 1, for the examination of risk of bias of each study [18]. All included studies presented detailed methodologies in terms of DNA extraction, NGS library preparation and sequencing, comparator typing protocols, and statistical analysis, with relevant scientific background and rationale explained. Only 3 included studies stated the eligibility criteria of participant selection [21- 23]. The 3 other studies obtained genomic samples from established sequencing databases or blood bank reference laboratories [24-26]. Data outcomes were presented in all 6 studies with an investigation on encountered discrepancies between NGS genotyping and alternative methods. Substantial evaluation of study limitations and potential confounders and bias were made in all studies, all clearly stated with their conflict of interest.
| Primary author; year | Scientific background and rationale explained | Eligibility criteria of participant selection described | Detailed study method illustrated | Outcome data reported | Limitations of the study discussed | Potential sources of bias addressed | Discrepant results investigated | Conflict of Interest Statement included |
|---|---|---|---|---|---|---|---|---|
| Fichou; 2016 [21] | Y | Y | Y | Y | Ya | Yb | n/ac | Y |
| Jakobsen; 2017 [22] | Y | Y | Y | Y | Yd | Ye | Y | Y |
| Orzinska; 2018 [23] | Y | Y | Y | Y | Yf | Yg | Y | Y |
| Boccoz; 2018 [24] | Y | n/ah | Y | Y | Yi | Yj | Y | Y |
| Paganini; 2020 [25] | Y | n/ah | Y | Y | Yk | Yl | Y | Y |
| Roulis; 2020 [26] | Y | n/ah | Y | Y | Ym, n | Ye | n/ac | Y |
Note: Y, criteria fulfilled; N, criteria not fulfilled.
aAlloimmunisation propensity beyond classification as weak or partial variants
bNGS data was retrospectively compared by Sanger sequencing and confirmed by phenotyping
cNo discrepant results to investigate
dNo full validation on NGS genotypes was available
eAnonymised data on ethnicities and determined genotypes of donors during the investigation
fInsufficient depth of sequence for some regions
gModification of primers suggested by software to ensure specific gene amplification
hGenomic samples retrieved from the formerly published databases or blood banks; recruitment period not applicable
iLow complexity and diversity of sequence in the experiment
jInvestigation on the relation between amplicon sizes and read counts considered due to the non-optimised size of amplicon before sequencing
kAmbiguous result reanalysis was not available
lSingular genetic mosaic profiles from geographic regions presented
mSample size discussed
nUnknown variants and limitations on probe design to target locus discussed
Table 1: Assessment checklist of risk of bias and quality of included studies, guided by STROBE checklist.
Characteristics of studies
The 6 eligible studies assessed are summarized with characteristics listed in Table 2 [21-26]. All studies employed a prospective study design in assessing the accuracy of NGS panels [22-25]. Samples were collected from either blood donors, or blood bank reference laboratory genomic DNA [21, 26]. All studies acquired samples from different ethnicities, including French, American, Danish, and Germany, Polish, European, Afghan, and Australian. Half of the 6 included studies used Thermo Fisher Ion Proton or Ion Torrent Pragmatic General Multicast (PGM) for sequencing, whilst in the other 3 studies, Ilumina MiSeq/NovaSeq sequencers were used for targeted NGS [21-26].
| Primary author; Year | Study design | Country | Sample size | Ethnicity | NGS platform | Comparator |
|---|---|---|---|---|---|---|
| Fichou; 2016 [21] | Prospective | France | 48 | French | Thermal Fisher Ion Proton or Ion PGM Sequencer | Sanger sequencing |
| Jakobsen; 2017 [22] | Prospective | U.S.A. | 72 | American, Danish, Germany | Thermal Fisher Ion PGM Hi-Q Sequencer | ID Core XT (Luminex-based assay) and Sanger Sequencing |
| Orzinska; 2018 [23] | Prospective | Poland | 45a | Polish | Thermal Fisher Ion Torrent PGM Sequencer | Real-time PCR (TaqMan) or commercially available test (FluoGene vERYfy) |
| Boccoz; 2018 [24] | Prospective | Europe | 95 | European | Illumina MiSeq nano flow cell sequencer | HEA BeadChip™ platform and HIFI Blood 96™, or serology |
| Paganini; 2020 [25] | Prospective | Afghanistan | 79 | Afghan | Illumina NovaSeq 6000 Sequencer | SNaPshot genotyping |
| Roulis; 2020 [26] | Prospective | Australia | 33a | Australian | Illumina MiSeq next generation sequencer | Real-time PCR (Taqman) |
| Note: a Study presented data of RBC, HPA, and/or HNA typing. Only data for RBC antigen genotyping was retrieved. | ||||||
Table 2: Characteristics of included studies in evaluating the concordance of red cell antigens genotyping by NGS.
The RBC genotyping results generated by NGS were analysed, and comparisons were made prospectively to different validation methods, including Sanger sequencing, serology, and an assortment of commercially available platforms, i.e., ID Core XT microarray, FluoGene vERYfy, HEA BeadChip array, SNaPshot, and TaqMan real-time PCR [21, 26]. In all studies but one, targeted whole genome sequencing was performed in 5 of the 6 included studies [21-25]. In the one remaining study, Whole Exome Sequencing (WGS) was adopted [26].
Concordance of NGS genotyping platforms
The concordance outcome of typing results between NGS and other comparators including serological and commercially available molecular typing methods was assessed for Kell, Kidd, and Duffy antigens separately by assessing the percentage agreement in genotyping results between NGS and the comparator, constituted the quantitative meta-analyses on each blood group.
In Kell genotyping, the agreement between individual studies before performing the arcsine transformation ranged from 91.8% to 100%, indicating a high degree of agreement. The pooled proportion agreement of 0.987 (95% CI, 0.975 to 0.996; P<0.001) was rated with statistical significance for the six included studies (Figure 2). The analysis reflected a clear absence of heterogeneity in the comparison, though with no statistical significance (I2=0%, P=0.805).
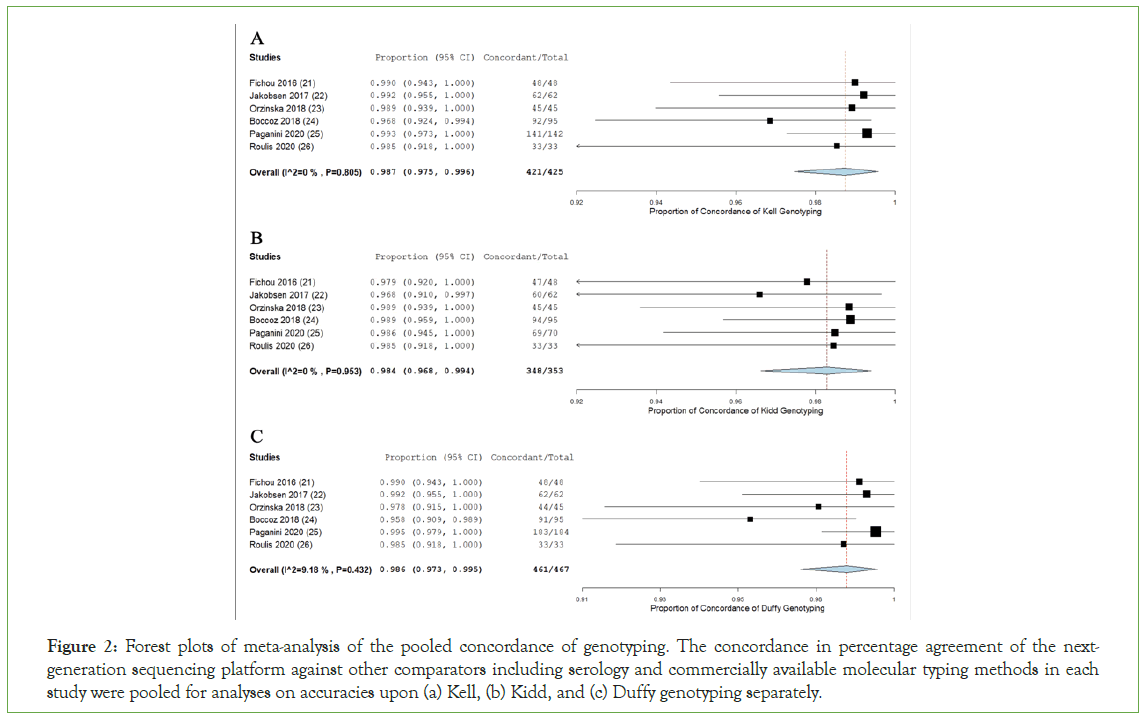
Figure 2: Forest plots of meta-analysis of the pooled concordance of genotyping. The concordance in percentage agreement of the nextgeneration sequencing platform against other comparators including serology and commercially available molecular typing methods in each study were pooled for analyses on accuracies upon (a) Kell, (b) Kidd, and (c) Duffy genotyping separately.
The agreement between individual studies before arcsine transformation ranged from 91.0% to 100%, indicating a high degree of agreement in Kidd genotyping. The pooled proportion agreement of 0.984 (95% CI, 0.968 to 0.994; P<0.001) was rated with statistical significance for the six included studies (Figure 2). No heterogeneity was observed, though not statistically significant (I2=0%, P=0.953).
For Duffy, the agreement between individual studies before arcsine transformation ranged from 95.8% to 100%, indicating a high degree of agreement. The pooled proportion agreement of 0.986 (95% CI, 0.973 to 0.995; P<0.001) was rated with statistical significance for the six included studies (Figure 3). No apparent heterogeneity was showed, with no statistical significance (I2=0%, P=0.432).
Figure 3: Heterogeneity of study or subgroup and statistical differences in overall test.
Depth of coverage for included data
The average depth of coverage varied between the range of 250X and 6474X within and across the included studies (Table 3). In Jakobsen et al., study, 2 out of the original 72 samples were omitted from analysis due to insufficient reads per amplicon [22]. Paganini et al., reported 35 unresolved events in total during WGS analysis attributed to low read counts corresponding to missing alleles and low read depth [25].
| Number of samples | Kell | Kidd | Duffy | ||||
|---|---|---|---|---|---|---|---|
| Mean depth coverage (reads) | Concordant results/samples compared | Mean depth coverage (reads) | Concordant results/samples compared | Mean depth coverage (reads) | Concordant results/samples compared | ||
| Fichou; 2016 [21] | 48 | 2,199 | 48/48 | 1,170 | 47/48 | 1,011 | 48/48 |
| Jakobsen; 2017 [22] | 72 | 2,389 | 62/62a | 1,428 | 60/62a | 1,058 | 62/62a |
| Orzinska; 2018 [23] | 45 | 5,122 | 45/45 | 6,474 | 45/45 | 5,806 | 44/45 |
| Boccoz; 2018 [24] | 95 | - b | 92/95 | - b | 94/95 | - b | 91/95 |
| Paganini; 2020 [25] | 79 | 1,445 | 141/142c | 2,637 | 69/70c | 552 | 183/84c |
| Roulis; 2020 [26] | 33 | ~250 | 33/33 | ~250 | 33/33 | ~250 | 33/33 |
Note: a Only data from 62 samples were genotyped using a comparator due to insufficient depth coverage, missing data, and error in data retrieval |
|||||||
Table 3: Summary of outcomes regarding mean depth coverage and concordance of Kell, Kidd, and Duffy genotyping.
Discrepancy in genotyping results
Discordant events were demonstrated in 5 of the 6 included studies, with only one study reporting 100% concordance agreement between NGS and the comparator methods (Table 4) [22-25]. In Fichou et al., study, one discrepant sample was reported with the expected Jk(a+wb+) phenotype by NGS disagreed with the known phenotype of Jk(a-b+). Jakobsen et al, reported two discrepant results with failure in JK*01/02 detection by NGS assay due to low read coverage, with true genotype confirmed by Sanger sequencing. One discrepancy was reported in Orzinska et al., study with the presence of a weak variant of Fyb antigen, FY*X causing disagreement between the donor phenotype of Fy (a+b-) and the detected FY*A and FY*B alleles. Boccoz et al., reported 8 discrepant events between platforms across blood groups attributing to low read counts, poor sample quality and errors on genes of interest in the early stages. Paganini et al., reported one incorrect typing in each blood group, which failed to predict the heterozygosity of samples.
| Study | Title and abstract provide summary and clear information | Introduction explains the scientific background | Introduction state specific objective Including any prespecified hypothesis | Presents key elements of study design | Describes the setting, relevant dates, and period of recruitment | Define all outcomes and exposures | Define any effort to address potential bias | Explain how study size was arrive | Gives diagnostic criteria | Summarise results And Discuss limitations |
|---|---|---|---|---|---|---|---|---|---|---|
| Vlachodimitropoulou 2021 | Y | Y | Y | Y | Y | Y | N | Y | Y | Y |
| Liu 2021 | Y | Y | Y | Y | Y | Y | Y | Y | Y | Ne |
| Ángeles 2019 | Y | Y | Y | Y | Y | Y | N | Y | Y | Y |
| Slootweg 2018 | Y | Y | Y | Y | Y | N | N | Y | Y | Y |
| Kamphuis 2007 | Y | N | Y | Y | Y | Y | Y | Y | Y | Y |
| Wamelen 2007 | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Mari 2000 | Y | Y | Y | N | N | Y | Y | N | Y | Ne |
| Rimon 2006 | Y | Y | Y | Y | Y | N | Y | Y | Y | Ne |
| Van Dongen 2006 | Y | Y | Y | Y | Y | Y | Y | Y | Y | N |
| Mckenna 1999 | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
Note: Y=yes, N=No, Ne=Limitation not discussed. |
||||||||||
Table 4: Assessment of included study’s methodological quality according to strobe checklist.
Capacity for detection of novel and rare variants
Five studies successfully detected rare variants in Kidd and Duffy among the samples analysed [21-23, 25, 26]. Two studies demonstrated the ability in detecting novel mutations [25, 26]. One SNP in the JK gene was identified in 37 samples in Paganini et al., study, Roulis et al., reported a potential novel P1PK null allele (c.202A>C) by NGS, though beyond the scope of blood group systems investigated in this research, presented the potential of NGS in the discovery of new blood group systems.
Discussion
NGS as a powerful alternative in red cell antigens typing
To assess the diagnostic performance of NGS in red cell antigen genotyping, Kell, Kidd, and Duffy in particular, this systematic review and meta-analysis was performed to summarize the published evidence on NGS typing concordance. Based on our results, it reveals that NGS has a high concordance of over 98% and is capable of being a screening tool with high accuracy in Kell, Kidd, and Duffy genotyping. While commercially available beads or microarray-based platforms may yield false-negative results from allele dropouts due to a lack of primer complementarity and interrogation on a limited number of SNPs, NGS offers a more comprehensive analysis by sequencing of whole genomes or exomes or by targeted strategies during massively parallel sequencing [27, 28]. Challenges in the identification of SNPs, indels, and structural variations during antigen prediction can be overcome by target enrichment NGS [28].
NGS goes beyond SNP detection encoding known alleles. Its strength allows for resolving complicated cases by simultaneous detection of multiple polymorphisms. In cohesion with the adequate mean depth coverage of genes mentioned in several publications, our included studies proved this strength in providing high-resolution interpretations of also rare and novel variants [21-23, 25, 26, 29-31]. Weinstock et al., reported a novel isoform p.42G (c.126 T>G) that is antithetical to the Fya- and Fyb-carrying proteins [32]. Fichou et al., study illustrated full concordance in defining the null alleles in Atypical Chemokine Receptor 1 (ACKR1) gene of the Duffy group among samples [21]. Three included studies also successfully reported the determination of weak antigens in our target blood groups [22, 23, 25]. Specifically, in Jakobsen et al., study, the weak Fyb antigens, encoded by FY*02M.01, in two donor samples were accurately resolved by NGS assay while conventional molecular comparator failed to determine genotype, manifesting the superiority of NGS in this context [22].
In addition to its role in blood group phenotype prediction, NGS platforms can resolve serological investigation upon blood typing discrepancies or ambiguities, such as the differentiation between partial D and weak D antigens, to guide clinical decision-making [27, 33]. Targeted exome sequencing superseded other molecular typing methods in resolving complicated cases from serology and SNV-typing as demonstrated by Schoeman et al., [34]. Also, the development of NGS provides a significant advance in personalized care management by favoring more research on minor alleles and their potential significance in the alloimmunisation [25].
Drawbacks in NGS implication in clinical practice
The obstacle is always around gene amplification and resolution. Primer design for NGS library preparation is challenging since pseudogenes and homologous genes with high sequence similarity pose difficulty in flanking target sequences, such as RHD/RHCE, and GYPA/GYPB genes, both with over 90% sequence identity, causing misalignment [6, 35, 36]. Genomic variation within the primer sites can also prompt allele dropout and thus incorrect typing. A special primer design or algorithm may be required for correct phasing. Besides, flanking regions with high GC content also affect amplification, failing in fully cover all coding regions [22].
Another challenge of NGS blood group genotyping is the complication concerning the prediction of clinical significance of previously undescribed genetic variations in the absence of clinical correlation. More importantly, high instrumental and sequencing costs as well as the cost of tremendous data storage, in conjunction with its high turnaround time of up to 56 hours impede NGS application in clinical settings [6, 37].
Future role of NGS in donor screening
Whilst high turnaround time is a common problem in NGS assays and therefore unfavorable to patient typing, this approach is undoubtedly an ideal tool for extensive donor typing, in which awaited time for test results poses less effect. It favors the collection and process of an optimal number of donor samples, combined with its capability of simultaneous investigation on abundant blood group alleles of individual donor [6]. A well-typed inventory of donor blood in blood banks can be expected in the future, therefore achieving the goal of matching prophylactic antigens, and reducing the risk of alloimmunisation in real-world clinical settings is crucial to transfusion-dependent patients.
To develop NGS as a standardized screening panel, the reference database needs to be supplemented with millions of genomic data and corresponding serological and genomic backgrounds to deal with numerous SNVs possibly detected by NGS [12]. Multiplexing of samples and additional blood group genes could be included in future studies.
Strengths and limitations of this review
Together with a focus on clinically significant blood groups and the coverage of different ethnicities in samples studied, this systematic review provides new insights into the accuracy of red cell antigen genotyping by NGS in favor of its use in clinical investigation. However, the initial search for eligible studies was limited to languages and accessibility, and to those in which comparisons were made for interested blood groups in terms of concordance despite four large databases employed. Moreover, the strength of results obtained is also influenced by the small sample size across studies, despite the high quality of individual studies with minimal risk of bias and confounding factors being validated.
Conclusion
The emergence of NGS platforms flavors an unprecedented contribution to massive donor screening and is predicted to play a solid role in securing transfusion safety and minimizing the risk of transfusion-related complications in patients. This systematic review/ meta-analysis accumulated evidence on employing NGS as a future technique in typing Kell, Kidd, and Duffy antigens whilst it provides a more in-depth insight into the genetic profile of donors and allows mass-scale screening with its extensive genotyping capability. Despite the concerns over its high running costs and requirement on data storage and management, this study proved the principle of high concordance blood group genotyping by NGS and its relevance in novel and rare allele’s investigation.
To date, limited literature has been published. Based on our observations, it implies the need of extending the investigation to larger pools of samples in conjunction with applying this technique to other blood group antigens genotyping to fill the gap between benchtop and clinical settings. Ideally, more technological advances in NGS platforms are also expected to achieve cost reduction to implement this approach in a clinical context in the future.
Conflict of Interest
None to report.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not for profit sectors.
References
- Red Cell Immunogenetics and Blood Group Terminology. ISBT. 2022.
- Smart E, Armstrong B, Lee E. Blood group systems. ISBT Sci Series. 2020;15(1):123-150.
- Dean L. Blood Groups and Red Cell Antigens. NCBI. 2005.
- Lee S, Zambas ED, Marsh WL, Redman CM. The human Kell blood group gene maps to chromosome 7q33 and its expression is restricted to erythroid cells. Blood. 1993; 81(10):2804-2809.
[Crossref] [Google Scholar] [PubMed]
- Guelsin GA, Sell AM, Castilho L, Masaki VL, de Melo FC, Hashimoto MN, et al. Genetic polymorphisms of Rh, Kell, Duffy and Kidd systems in a population from the State of Parana, southern Brazil. Rev Bras Hematol Hemoter. 2011;33(1):21-25.
[Crossref] [Google Scholar] [PubMed]
- Furst D, Tsamadou C, Neuchel C, Schrezenmeier H, Mytilineos J, Weinstock C, et al. Next-Generation Sequencing Technologies in Blood Group Typing. Transfus Med Hemother. 2020;47(1):4-13.
[Crossref] [Google Scholar] [PubMed]
- Daniels G, Poole J, De Silva M, Callaghan T, MacLennan S, Smith N, et al. The clinical significance of blood group antibodies. Transfus Med. 2002;12(5):287-295.
[Crossref] [Google Scholar] [PubMed]
- Kleinman S, Chan P, Robillard P. Risks associated with transfusion of cellular blood components in Canada. Transfus Med Rev. 2003;17(2):120-162.
[Crossref] [Google Scholar] [PubMed]
- Mitra R, Mishra N, Rath GP. Blood groups systems. Indian J Anaesth. 2014;58(5):524-528.
[Crossref] [Google Scholar] [PubMed]
- Moulds J. An Overview of the Classic Serological Methods: Limitations and Benefits of Serology and DNA Testing. NY: Sprin New York. 2010; 54:1-7.
- Quraishy N, Sapatnekar S. Advances in Blood Typing. Adv Clin Chem. 2016; 221-269.
[Crossref] [Google Scholar] [PubMed]
- Orzinska A, Guz K, Brojer E. Potential of next-generation sequencing to match blood group antigens for transfusion. Int J Clin Trans Med. 2019; 7:11-22.
- Quirino MG, Colli CM, Macedo LC, Sell AM, Visentainer JE. Methods for blood group antigens detection: cost-effectiveness analysis of phenotyping and genotyping. Hematol Transfus Cell Ther. 2019; 41(1):44-49.
[Crossref] [Google Scholar] [PubMed]
- Krishna B M, Khan M, Khan S. Next-Generation Sequencing (NGS) Platforms: An Exciting Era of Genome Sequence Analysis. Microb Gen Sus Agro Eco. 2019;2:89-109.
- Westhoff CM, Reid ME. Review: the Kell, Duffy, and Kidd blood group systems. Immunohematology. 2004;20(1):37-49.
[Crossref] [Google Scholar] [PubMed]
- Castilho L, Rios M, Pellegrino JJ, Carvalho MH, Alberto FL, Saad ST, et al. Genotyping of Kell, Duffy, Kidd and RHD in patients with b Thalassemia. Rev Bras Hematol Hemoter. 2000;22(2):69-76.
- von Elm E, Altman DG, Egger M, Pocock SJ, Gotzsche PC, Vandenbroucke JP, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. J Clin Epidemiol. 2008;61(4):344-349.
[Crossref] [Google Scholar] [PubMed]
- Wallace BC, Dahabreh IJ, Trikalinos TA, Lau J, Trow P, Schmid CH, et al. Closing the Gap between Methodologists and End-Users: R as a Computational Back-End. J Stat Software. 2012; 49(5):1-15.
- Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557-560.
[Crossref] [Google Scholar] [PubMed]
- Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gotzsche PC, Ioannidis JP, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339:2700.
[Crossref] [Google Scholar] [PubMed]
- Fichou Y, Mariez M, Le Marechal C, Ferec C. The experience of extended blood group genotyping by next-generation sequencing (NGS): investigation of patients with sickle-cell disease. Vox Sang. 2016;111(4):418-424.
[Crossref] [Google Scholar] [PubMed]
- Jakobsen MA, Dellgren C, Sheppard C, Yazer M, Sprogoe U. The use of next-generation sequencing for the determination of rare blood group genotypes. Transfus Med. 2019;29(3):162-168.
[Crossref] [Google Scholar] [PubMed]
- Orzinska A, Guz K, Mikula M, Kulecka M, Kluska A, Balabas A, et al. A preliminary evaluation of next-generation sequencing as a screening tool for targeted genotyping of erythrocyte and platelet antigens in blood donors. Blood Transfus. 2018;16(3):285-292.
[Crossref] [Google Scholar] [PubMed]
- Boccoz SA, Fouret J, Roche M, Lachuer J, Legras-Lachuer C, Corgier BP, et al. Massively parallel and multiplex blood group genotyping using next-generation-sequencing. Clin Biochem. 2018;60:71-76.
[Crossref] [Google Scholar] [PubMed]
- Paganini J, Nagy PL, Rouse N, Gouret P, Chiaroni J, Picard C, et al. Blood group typing from whole-genome sequencing data. PLOS ONE. 2020;15(11):2421-2468.
[Crossref] [Google Scholar] [PubMed]
- Roulis E, Schoeman E, Hobbs M, Jones G, Burton M, Pahn G, et al. Targeted exome sequencing designed for blood group, platelet, and neutrophil antigen investigations: Proof-of-principle study for a customized single-test system. Transfusion. 2020;60(9):2108-2120.
[Crossref] [Google Scholar] [PubMed]
- Westhoff CM. Blood group genotyping. Blood. 2019;133(17):1814-1820.
[Crossref] [Google Scholar] [PubMed]
- Liu Z, Liu M, Mercado T, Illoh O, Davey R. Extended Blood Group Molecular Typing and Next-Generation Sequencing. Transfus Med Rev. 2014;28(4):177-186.
[Crossref] [Google Scholar] [PubMed]
- Lane WJ, Westhoff CM, Uy JM, Aguad M, Smeland-Wagman R, Kaufman RM, et al. Comprehensive red blood cell and platelet antigen prediction from whole genome sequencing: Proof of principle. Transfusion. 2016;56(3):743-754.
[Crossref] [Google Scholar] [PubMed]
- Moller M, Joud M, Storry JR, Olsson ML. Erythrogene: a database for in-depth analysis of the extensive variation in 36 blood group systems in the 1000 Genomes Project. Blood Adv. 2016;1(3):240-249.
[Crossref] [Google Scholar] [PubMed]
- Schoeman EM, Lopez GH, McGowan EC, Millard GM, O'Brien H, Roulis EV, et al. Evaluation of targeted exome sequencing for 28 protein-based blood group systems, including the homologous gene systems, for blood group genotyping. Transfusion. 2017;57(4):1078-1088.
[Crossref] [Google Scholar] [PubMed]
- Weinstock C, Mytilineos J, Bugert P, Sitzmann N, Pensel E, Schrezenmeier H, et al. A novel allele of the atypical chemokine receptor 1 (ACKR1) gene containing the nucleotide change c.126 T>G (p.42Glu) encodes a third Duffy blood group protein sequence antithetical to that encoding Fya and Fyb antigens. Transfusion. 2019;59(6):2158-2159.
[Crossref] [Google Scholar] [PubMed]
- Castilho L. Molecular typing of blood group genes in diagnostics. Ann Blood. 2021;6:20.
- Schoeman EM, Roulis EV, Liew YW, Martin JR, Powley T, Wilson B, et al. Targeted exome sequencing defines novel and rare variants in complex blood group serology cases for a red blood cell reference laboratory setting. Transfusion. 2018;58(2):284-293.
[Crossref] [Google Scholar] [PubMed]
- Levan KC, Mouro I, Cherif-Zahar B, Raynal V, Cherrier C, Cartron JP, et al. Molecular cloning and primary structure of the human blood group RhD polypeptide. Proc Natl Acad Sci U S A. 1992;89(22):10925-10929.
[Crossref] [Google Scholar] [PubMed]
- Kudo S, Fukuda M. Structural organization of glycophorin A and B genes: glycophorin B gene evolved by homologous recombination at Alu repeat sequences. Proc Natl Acad Sci U S A. 1989;86(12):4619-4623.
[Crossref] [Google Scholar] [PubMed]
- Krumm N, Hoffman N. Practical estimation of cloud storage costs for clinical genomic data. Pract Lab Med. 2020;21:16-18.
[Crossref] [Google Scholar] [PubMed]
Citation: Hua HC, Jackson DE (2023) Next Generation Sequencing of Human Red Cell Antigens Kell, Kidd, and Duffy for Routine Clinical Investigations and Donor Screening: A Systematic Review and Meta-Analysis. J Blood Disord Transfus. 14:552.
Copyright: © 2023 Hua HC, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.