Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- CiteFactor
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research - (2020) Volume 11, Issue 4
Molecular identification of Jujube witches'-broom phytoplasma (16SrV) associated with witches'-broom disease of Ziziphus oenoplia in India
Snehi SK*, Srivastava S, Parihar SS and Jain BReceived: 06-Jan-2020 Published: 16-Apr-2020, DOI: 10.35248/2157-7471.20.11.492
Abstract
Severe witches’-broom disease of Ziziphus oenoplia was observed with significant disease incidence in Bhopal, India, during 2019. Phytoplasma was detected from symptomatic leaf samples by polymerase chain reaction (PCR) using phytoplasma 16S rRNA gene specific primers which revealed positive amplification of expected size ~1.2 kb DNA band. The positive amplicons of the phytoplasma 16S rRNA (1.2 kb) were sequence and sequenced data was submitted in GenBank database (Accession no. MK975463 and MK975462). On the basis of highest 99% sequence identities, closest phylogenetic relationships and In silico of the under study both the phytoplasma isolates associated with witches'-broom disease of Ziziphus oenoplia identified as a species of Jujube witches'-broom phytoplasma as a member of Elm yellows group (16SrV). To the best of our knowledge, this is the first report on the association of Jujube witches'-broom phytoplasma species of Elm yellows group (16SrV) with witches'-broom disease of Z. oenoplia in India.
Keywords
Ziziphus oenoplia; PCR; Sequence analyses; Jujube witches'-broom phytoplasma
Introduction
Phytoplasmas are intracellular obligate prokaryotes which lack cell wall, have small genome and are mainly transmitted by hemipteran insect vector of the families Cicadellidea (leafhoppers) and Fulgoridea (planthopper) [1]. Phytoplasma are associated with typical phyllody, virescence, yellowing, proliferation of axillary buds, witches’ broom, stunting of whole plant and die back symptoms on number of plant species worldwide [2,3]. Phytoplasma are also associated with severe yield losses in a variety of plant species of horticultural, agricultural and ornamental importance in India [4].
Ziziphus oenoplia (L.) Mill. (Family Rhamnaceae) commonly well known as makai in hindi and Jackal Jujube in english, is a straggling shrub distributed all over the hotter regions of Pakistan, Sri Lanka, India, Malaysia, and Tropical Asia [5]. The fruits are edible and it is widely used in Ayurveda for the treatment of various diseases, such as ulcer, stomach ache, obesity, asthma, digestive, antiseptic, hepatoprotective, wound healing and diuretic property [6].
There are limited reports available in the literature worldwide related to phytoplasma study in Ziziphus species such as ‘Candidatus Phytoplasma ziziphi’, associated with Jujube witches’ broom in China, Japan and Korea [7]. Phytoplasmas associated with witches’-broom disease in Ziziphus jujube and Z. nummularia in Bahraich district, in India, are considered isolates of ‘Ca. Phytoplasma ziziphi ’ [8]. Presently only one report has been published based on symptomatology on Ziziphus oenoplia expressed witches’-broom appearance by proliferation of axillary buds from Dakshin Dinajpur district of West Bengal, India West Bangal [9].
Barkatullah University (BU) campus, Bhopal is rich from plant diversity and various plant species are grown naturally and one of them some plant species may be naturally associated with some phytopathogens. At present no report is available of phytoplasma disease on Ziziphus oenoplia from India, therefore, molecular identification of phytoplasma naturally occurring on Ziziphus oenoplia grown in Bhopal was carried out in this study.
Materials and Methods
Symptomatology and survey
The Z. oenoplia plants were found to phytoplasma like symptoms rosetting, proliferation of axillary shoots, exhibiting witches ’ - broom, excessive branching accompanied with little leaf symptoms with 40-45% disease incidence during the survey in February, 2019 in Barkatullah University campus, Bhopal (Figure 1).

Figure 1. Symptoms showing witches- broom (a & c) compared with healthy (b & d) of Ziziphus oenoplia growing naturally at Barkatullah Univeristy, Bhopal.
DNA extraction
For molecular detection of phytoplasma total DNA was isolated from symptomatic and asymptomatic plant leave samples (100 mg) using phytoplasma enrichment protocol and quantity of DNA preparation as checked by taking its O.D. at 260/280 nm is 1.8 and concentration is also checked by the 1% agarose gel electrophoresis.
Polymerase chain reaction (PCR) and nested PCR
The phytoplasma 16S rRNA gene was detected by direct PCR using P1/P6 primers [10] and nested PCR carried out with universal primers pair R16F2n/R16R2[11] employing the PCR (P1/P6) product as a template DNA (1: 10) resulted in expected size positive amplification ~1.2 kb in symptomatic (4/4) leaf samples and not in asymptomatic healthy (1/1) leaf sample (Figure 2). The nested PCR conditions were: denaturation at 94°C for 5 min, followed by 30 cycles of 94°C for 50 s, 55°C for 45 s and 72°C for 90 s and a final extension for 7 min at 72°C.
The two positive nested PCR amplicons (~1.2 kb) were purified by Wizard SV gel extraction kit (Promega Pvt., Ltd., USA) and sequenced from both the direction (Bioinovations Pvt. Ltd., Mumbai, India). The ~1.2 kb sequence data of partial 16S rRNA gene were analyses and identical sequence data submitted in NCBI GenBank database under accessions: MK975463 and MK975462 (Figure 2).
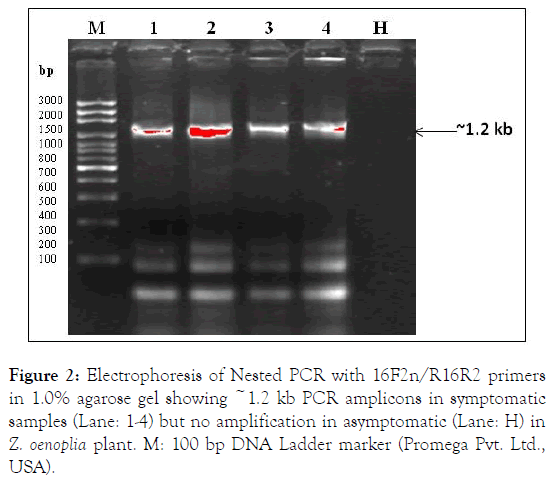
Figure 2. Electrophoresis of Nested PCR with 16F2n/R16R2 primers in 1.0% agarose gel showing ~1.2 kb PCR amplicons in symptomatic samples (Lane: 1-4) but no amplification in asymptomatic (Lane: H) in Z. oenoplia plant. M: 100 bp DNA Ladder marker (Promega Pvt. Ltd., USA).
Computational analysis of sequence data
The sequence data obtained through sequencing results was analyzed for consensus data remaining no ambiguities and submitted in National Centre for Biotechnology Information GenBank database (NCBI). To observe the nucleotide identity within and with other reported strains of phytoplasma, basic local alignment search tool (BLAST) searches were performed with all available databases using the NCBI-BLAST server.
Phylogenetic analyses were perused using Molecular Evolutionary Genetics Analysis (MEGA version 7.1) program with 100 replicates bootstrapping and phylogram were generated with Neighbour-joining method. Dendrograms were viewed by the NJplot program. In silico RFLP analysis of 16S rRNA sequences of phytoplasmas strain under study were generated using pDRAW32 program.
Results and Discussion
The natural occurrence of witches’- broom with little leaf disease of Z. oenoplia was detected by nested PCR using phytoplasma specific primers and 1.2 kb sequence data were analysed by NCBI BLASTn.
BLASTn analysis of 16S rRNA gene of understudy both the isolates (MK975463 and MK975462) showed highest 99% sequence identities with Jujube witches'-broom phytoplasma (MH972556, MH972553, MH972548) of Ziziphus sap sucking insects from India and ‘ Candidatus Phytoplama balanitae ’ (HG937644, LT558785, MH819290) of Elm yellows group (16SrV) of Ziziphus oenoplia and Zizyphus mauritiana from India. During phylogenetic analysis (MEGA v 7.1) of under study both the isolates (MK975463 and MK975462) shared closest relationships with the phytoplasma species of Jujube witches'- broom phytoplasma (MH972556, H744152 & MH972548) and ‘ Ca. Phytoplama balanitae ’ (LT558785) a member of Elm yellows group (16SrV) (Figure 3).
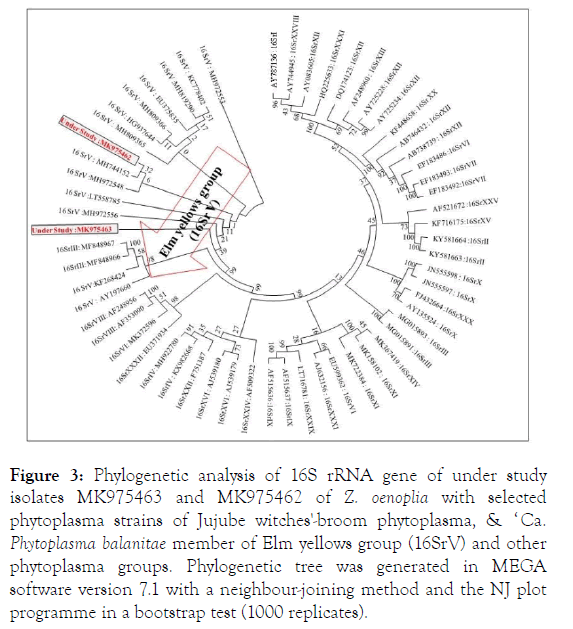
Figure 3. Phylogenetic analysis of 16S rRNA gene of under study isolates MK975463 and MK975462 of Z. oenoplia with selected phytoplasma strains of Jujube witches'-broom phytoplasma, & ‘ Ca. Phytoplasma balanitae member of Elm yellows group (16SrV) and other phytoplasma groups. Phylogenetic tree was generated in MEGA software version 7.1 with a neighbour-joining method and the NJ plot programme in a bootstrap test (1000 replicates).
The status of phytoplasma strain under study (MK975463 and MK975462) were also verified by In silico RFLP analysis with isolates of Jujube witches'-broom phytoplasma (MH972556) and ‘ Ca. Phytoplasma balanitae ’ (HG937644) from India of Elm yellows group (16SrV) using 06 restriction enzymes. In silico RFLP analysis of 16S rRNA sequences of phytoplasmas strain under study were generated using pDRAW32 program. Each 16S rRNA sequences were digested In silico with restriction enzymes: AluI, BamHI, EcoRI, HaeIII, KpnI, and TaqI and a virtual gel electrophoresis image were generated. The analysis In silico RFLP revealed a silently difference between the Elm yellows group (16SrV) taken for study. RFLP analysis with BamHI and KpnI showed no sites in phytoplasma strain both the under study isolates (MK975463 and MK975462) as well as any of the Jujube witches'-broom phytoplasma (MH972556) and ‘ Ca. P. balanitae’ (HG937644) 16SrVI group representative (Table 1).
| Enzymes | MK975463 | MK975462 | MH972556 | HG937644 |
|---|---|---|---|---|
| (Under study) | (Under study) | (Jujube witches'-broom phytoplasma) | (‘Ca. Phytoplasma balanitae) | |
| AluI | 706, 247, 145, 77, 45 | 706, 247, 147, 77, 45 | 706, 247, 145, 73 | 706, 271, 277, 247, 132, 77 |
| BamHI | - | - | - | - |
| EcoRI | 696, 524 | 696, 526 | 726, 524 | 783, 650 |
| HaeIII | 894, 188, 138 | 894, 190,138 | 894, 188, 168 | 894, 314, 225 |
| KpnI | - | - | - | - |
| TaqI | 804, 358,58 | 806, 358, 58 | 804, 358, 88 | 888, 358, 145, 42 |
Table 1: In silico analysis of 16S rRNA of phytoplasma under study (MK975463 and MK975462) along with other of Jujube witches'-broom phytoplasma (MH972556) and ‘Candidatus Phytoplasma balanitae’ (HG937644) member of Elm yellows group (16SrV) from India using 6 selected restriction enzymes (band length in bp).
The In silico restriction digestions and virtural gel plotting also suggested that phytoplasma both the under study isolates (MK975463 and MK975462) associated with witches-broom of is a species of Jujube witches'-broom phytoplasma (MH972556) and ‘Ca. P. balanitae’ (HG937644) of Elm yellows group (16SrV) (Figure 4).
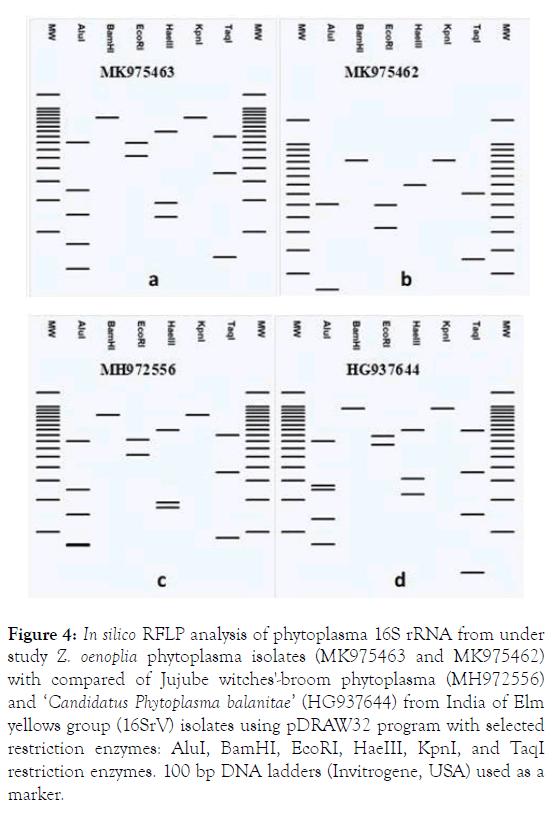
Figure 4. In silico RFLP analysis of phytoplasma 16S rRNA from under study Z. oenoplia phytoplasma isolates (MK975463 and MK975462) with compared of Jujube witches'-broom phytoplasma (MH972556) and ‘Candidatus Phytoplasma balanitae’ (HG937644) from India of Elm yellows group (16SrV) isolates using pDRAW32 program with selected restriction enzymes: AluI, BamHI, EcoRI, HaeIII, KpnI, and TaqI restriction enzymes. 100 bp DNA ladders (Invitrogene, USA) used as a marker.
There are limited reports available in the literature worldwide related to phytoplasma study in Ziziphus species such as ‘Ca. Phytoplasma ziziphi’ in China, Japan, Korea and India [7,8]. The Z. oenoplia is totally different in their nature to other Ziziphus plant species. It is basically a creeper plant and spreading, sometimes climbing, however the possibility of phytoplasma infection is more prominent to spread the phytoplasma disease to one healthy plant species to another healthy plant species. Therefore detection and identification of phytoplasma species is more essential for proper management of phytoplasma disease.
Conclusion
On the basis of sequence analysis, closest phylogenetic relationships and In silico RFLP of the under study both the phytoplasma isolates associated with witches'-broom disease of Ziziphus oenoplia identified as a strain of Jujube witches'-broom phytoplasma as a member of Elm yellows group (16SrV ) from Barkatullah University, Bhopal, Madhya Pradesh, India. To the best of our knowledge, this is the first report on the association of Jujube witches'-broom phytoplasma species of Elm yellows group (16SrV) with witches'-broom disease of Z. oenoplia in India.
Acknowledgments
The authors would like to thank to Head Department of Microbiology and Vice chancellor, Barkatullah University, Bhopal for support and encouragement.
REFERENCES
- Lee IM, Davis RE,Gundersen-Rindal DE. Phytoplasma: Phytopathogenic Mollicutes. AnnRev Micro. 2000;32:221-225.
- Al-Saady NA, Khan AJ. Phytoplasmas that can infect diverse plant species worldwide. Phy Mol Biol Plants. 2006;12:263-281.
- Raj SK, Snehi SK, Khan MS, Singh M, Chaturvedi Y, Tiwari AK, et al. Molecular detection and identification of phytoplasma isolates associated with little leaf and witches'-broom disease of marigold (Tageteserecta L.) in India. Phytopath Mollicutes. 2011;1:41-46.
- Chaturvedi Y, Rao GP, Tewari AK, Duduk B, Bertaccini A. Phytoplasma on ornamentals: Detection, diversity and management. Acta Phytopath et Entomol Hungarica. 2010;45:31-69.
- Pullaiah T. Medicinal Plants in Andhra Pradesh. Illustrated (2nd edn). Daya books. 2002;406-407.
- Rao V, Rawat AKS, Singh AP. Hepato protective potential of Ethanolic extract of (L) Mill roots against Ziziphus oenoplia antitubercular drugs induced hepato toxicity in experimental models. Asian Pacific J Trop Medicine. 2012;283-288.
- Jung HY, Sawayanagi T, Kakizawa S. ‘Candidatus Phytoplasma castaneae’, a novel phytoplasma taxon associated with chestnut witches’ broom disease. Int J System Evol Micro. 2003;53:1037–1041.
- Khan MS, Raj SK, SnehiSK. Natural occurrence of ‘Candidatus Phytoplasma ziziphi’ isolates in two species of jujube trees (Ziziphus spp.) in India. Plant Pathol. 2008;57:1173.
- Chakraborty KT. Notes on phytoplasma diseases from Dakshin Dinajpur district of West Bengal, India. Ann Plant Sci. 2017;6:1866-1867.
- Deng S, Hiruki D. Amplification of 16S rRNA genes from culturable and nonculturable mollicutes. J Micro Methods. 1991;14:53-61.
- Gundersen DE, Lee IM. Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopathol Mediterranea. 1996;35:144-151.
Citation: Snehi SK, Srivastava S, Parihar SS, Jain B (2020) Molecular Identification of Jujube Witches‘-Broom Phytoplasma (16srv) Associated With Witches’-Broom Disease of Ziziphus oenoplia in India. Plant Pathol Microbiol. 11:492. doi: 10.35248/2157-7471.20.11.492.
Copyright: © 2020 Snehi SK, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

