Indexed In
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- China National Knowledge Infrastructure (CNKI)
- Electronic Journals Library
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Editorial - (2012) Volume 3, Issue 5
Kinetic Modeling of Microbiological Processes
Kinetic description of microbiological processes is vital for the design and control of microbe-based biotechnologies such as waste water treatment, petroleum oil recovery, and contaminant attenuation and remediation. Various models have been proposed to describe microbiological processes. Monod equation, which is named for Noble Prize-winning French biologist Jacques Monod, is by far the most widely used approach. The model relates microbial growth rates to the concentration of a limiting nutrient, for example, dissolved organic carbon, using a hyperbolic equation. The Monod equation has the same form as the Michaelis-Menten equation for enzymatic kinetics. However, the former is considered to be empirical, while the latter has been proved theoretically. Whether the Monod kinetics is theoretically correct, at least macroscopically, remains to be found. We may recall that the theory of the Michaelis-Menten enzymatic kinetics involves the formation of an enzyme-substrate complex as the key hypothetical step to hyperbolically link the rate and substrate concentration. This hypothesis was not proved until many years later when the existence of such a complex was experimentally demonstrated.
Various derivative versions of the Monod model have been proposed to describe other limiting conditions, for example, dual or multiple Monod model to account for the simultaneous limitation of other substrates and nutrients. One important finding in the application of the Monod-type models is the limitation of reaction free energy for bacterial respiration in energy-limited environments. Unlike chemical reaction kinetics where a forward reaction does not exhaust its driving force until its reaction free energy becomes zero, a microbiological reaction ceases when the gain of reaction free energy in the respiration is less than a certain value, which is sometimes termed as the minimum free energy. The minimum free energy relates to the production of intracellular molecule ATP, an energy storage biochemical in living cells. The magnitude and origin of the minimum free energy is, however, interpreted differently. One interpretation is arrived from energetic analysis of biochemical reactions at the substrate level that the overall respiration reaction must generate enough energy to synthesize at least one ATP (31.8 kJ/mol under the standard state chemical condition, and approximately 50 kJ/mol under typical intracellular conditions). Energetic analysis of microbial community structure as well as the existence of microorganisms in energy-deficient environments, however, indicated that the minimum energy required to sustain microbiological activities is much less, approximately equivalent to generate a third ATP. An alternative interpretation is to attribute the minimum free energy to the energy required to eject one proton from cytoplasm to periplasm to form membrane-associated proton motive force, which is another form of cell energy that can be exploited to generate ATP and drive other intracellular reactions and transport processes. Because each ATP production requires 3 protons, the average free energy required to eject one proton is a third of the ATP formation. The mechanistic details for the linkage between the proton motive force and energy limitation for microbiological processes have not been worked out.
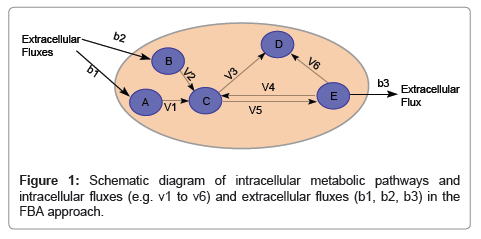
One major shortcoming of the Monod and its derivative models is that it cannot describe dynamic physiological response to changing environments. The activity (physiology) of microorganisms depends on their interactions with other microbes and the environment (ecology). At the intracellular level, many alternative metabolic pathways as encoded in their genome can be activated or deactivated depending on extracellular environment. An approach to account for the impact of potential switching between these alternative pathways to the rates of microbiological processes is the constraint-based models. The approach intends to reconstruct metabolic pathways by identifying, categorizing, and interconnecting genes, proteins, reactions and metabolites that participate in the metabolic activity of a biological system to form a reaction network that describes fundamental metabolic pathways. Rigorous application of the reaction network for modeling microbiological processes is, however, currently infeasible because of the challenges in characterizing the kinetic properties for each reaction in the network, which typically contains hundreds of reactions. There are two approximating approaches to circumvent this difficulty in implementing the constraint-based approach. One is to reduce the number of reactions in the reaction network until it becomes feasible to experimentally characterize the kinetics of remaining reactions. Another is the Flux Balance Approach (FBA) by assuming a rapid kinetics of each intracellular reaction relative to the changes of extracellular processes so that a steady-state flux balance equation can be applied for each reaction (Figure 1). The steady-state flux equations for different intracellular reactions are then linked based on the mass balance to form a set of linear equations, which can be readily solved using linear programming to determine steady-state reaction flux distribution in a metabolic network by maximizing an objective function, such as ATP production or growth rate. The FBA approach has received more attention than the kinetic approach with a reduced number of reactions.
The FBA has been applied in the fields of system biology,bioinformatics, genomics, and bioproduct discovery. The application of the approach in petroleum and environmental biotechnologies will require extensive computer resources for optimization process, especially under conditions when external conditions change dynamically such as in subsurface porous media. Another limitation of the approach is the availability of metabolic pathways and reaction networks that have only been determined for a limited number of bacteria. This limitation will, however, relieve with the rapid advancing of characterizing approaches such as high-throughput sequencing, high-density microarrays and proteomics. On the other hand, increasing biological reaction networks in the modeling will exponentially increase computational burden. Nevertheless, the major limitation of the FBA approach is that it ignores species concentrations, mass actions of species in the reaction network, and reaction energetics. The optimal metabolic pathways determined from the FBA can violate thermodynamic feasibility and/or kinetic energy barrier constraints. Research has been performed to address thermodynamic constraint by adding energy balance equations into the FBA approach by treating each reaction free energy as additional variable in the linear programming and by imposing energy flux direction to constrain thermodynamic feasibility. The modified approach is termed as the Energy Balance Approach (EBA). It is unclear, however, whether the reaction free energies optimized from the linear programming are consistent with the true chemical reaction free energies, which, by definition, are a function of species concentrations that are not considered in FBA/ EBA. The kinetic energy barrier constraint has not been investigated.
Mixed Monod-based and constraint-based approaches might offer the advantage in modeling microbiological processes in petroleum and environmental biotechnologies. Ideally, the EBA is used to provide optimal, thermodynamically feasible metabolic pathways that feed into the calculation of parameters in the Monod-type models while the Monod-types models service as the upscaled form to describe macroscopic manifestation of intracellular processes under dynamic environmental conditions and to provide extracellular fluxes that can be used to optimize metabolic pathways. Researchers are, however, needed to mathematically link these two types of models.
Copyright: © 2012 Liu C, et al. This is an open-access article distributed underthe terms of the Creative Commons Attribution License, which permits unrestricteduse, distribution, and reproduction in any medium, provided the original author andsource are credited.