Indexed In
- Academic Journals Database
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- China National Knowledge Infrastructure (CNKI)
- Scimago
- Ulrich's Periodicals Directory
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- MIAR
- University Grants Commission
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Review Article - (2022) Volume 13, Issue 2
Isolation, Identification and Molecular Characterization of Pasteurella multocida from Cases of Cattle from Selected Areas
Jaleta Shuka*, Shimelis Tesfaye, Bilisuma Ababae and Takele AbayinehReceived: 13-Apr-2022, Manuscript No. JVV-22-16114; Editor assigned: 15-Apr-2022, Pre QC No. JVV-22-16114 (PQ); Reviewed: 06-May-2022, QC No. JVV-22-16114; Revised: 13-May-2022, Manuscript No. JVV-22-16114 (R); Published: 20-May-2022, DOI: 10.35248/2157-7560.21.13.478
Abstract
The current research work was conducted from November 2019 to August 2020 to isolate, identify and molecularly characterizing Pasteurella multocida from cattle diseased with Hemorrhagic Septicemia (HS) in Bambasi and Assosa districts in Benishangul Gumuz Regional State with the objective of the existing vaccine improvement against Bovine pasturellois diseases in Ethiopia. For this purpose, a total of 30 samples were collected from clinical cases following outbreaks of HS in cattle. The samples were transported to National Veterinary Institute of Ethiopia using transport medium and then processed for isolation of P. Multocida. There were 4 samples isolated as P. Multocida. The isolates were identified by cultural and morphological features, biochemical features and pathogenicity tests. With the aim of additional confirmation by means of Pathogenicity study and inocucity estimation, the 4 isolates were inoculated on rabbits and the 4 (100%) isolates were found to be confirmed as Pasteurella up on post-mortem examination. The study of PCR assay revealed the presence of P. Multocida serotype B2, and absence of P. Multocida serotype E2. In conclusions P. Multocida serotype B2 in the designated area is considered as possible pathogens in resulting HS in cattle. However, further study supported by DNA sequencing is essential to finalize these conclusions. At the end based on the conclusions made, the recommendations were forwarded.
Keywords
Asosa; Bambasi; Cattle; Haemorrhagic; septicemia; Pasteurella multocida; Polymerase chain reaction
Abbrevations
ALFD: Assosa District Office of Livestock and Fishery Development; BLFD: Bambasi District office of Livestock and Fishery Development; CLSI: Clinical and lessons Standard Institute; CSA: Central Statistical Agency; CSY: Casein Sucrose Yeast; EA: Environmental Agency; FMD: Foot and Mouth Disease; HS: Hemorrhagic Septicemia; NMSA: National Meteorology Service Agency; NVI: National Veterinary Institute; OMA: Outer Membrane Protein; SDS: Sodium Dodecyl Sulfate; TEM: Transport Enrichment Medium.
Introduction
Even though Ethiopia is one of the largest countries with cattle population in Africa the productivity from the livestock sector has been insignificant due to the existence of infectious diseases like Pasteurellosis which affect the productivity of the sector [1,2].
There are two forms of pasteurellosis namely; Pneumonic and Septicemic pasteurellosis. Septicemic pasteurellosis is caused by P. Multocida. It is supposed that Louis Pasteur was a person who identified for the first time the bacterium P. Multocida as the causative agent of fowl cholera before 125 years ago. During his essential experiment he also showed though many passage this bacterium loses its virulence activity, but still inoculation the attenuated strain can enhance the protective immune response [3].
The species name “Multocida”in Latin means killing several which was invented viewing predominantly considerable mortality related as a consequence infection by this bacterium [4]. Among the family of pasturelliace P. Multocida consists of a varied group of gram negative bacteria which are liable for wide range of economically important infections of animals, including cattle and buffalo [5].
Morphologically, P. Multocida comprises a small, gram-negative, non-spore-forming, none-motile and penicillin sensitive coccobacillus with bipolar staining characters. The bacteria characteristically seem as a single bacillus on Grams’ stain: though, pairs and short chains can also be seen [6]. Based on serological classification the strains of P. multocida are currently grouped in to five groups namely; A, B D E, and F according to capsular configurations. Similarly, based on somatic serotype classification it is grouped in to 16 somatic serovars (1-16).
Based on serological classification the strains of P. Multocida are currently grouped in to five groups namely; A, B, D, E, and F capsular sero groups. Similarly, based on somatic serotype classification it is grouped in to 16 somatic serovars (1-16). It can also exist assembled based on their outer membrane protein type 16S rRNA-type. The ribosomal RNA (r RNA) of gram negative bacteria consists of two structures namely; 16 rRNA and 23 rRNA. Thus based on the 16 rRNA-classifications there are seven 16S type which belongs to a single family of the majority of clinical [7].
P. Multocida frequently occurs as a commensal in the upper respiratory tracts of several live stocks, poultry and domestic pet’s species [8]. However, it is kown to cause a wide range of economically important diseases like Haemorrhagic Septicemia (HS) in hooved animals specifically in cattle and buffalo in many Asian and African countries resulting in to high death and morbidity [9-11].The stains of P. Multocida causing HS consists of serotype B:2, B: 2, 5 and B;5 in Asian countries and E;2 in African countries [12]. Moreover, in addition to causing HS it is known to result I atrophic rhinitis in Swine, fowl cholera in poultry [4,13]. Serogroup A and D are identified to infect calves, pigs,’ sheep, goats and rabbits in which they cause different infections [14]. Moreover, serogroup F has been identified as a causal agent of fowl cholera but [15], in latest period, it has similarly been recognized in some mammalian species in different biosphere parts [16].
HS is a very acute, lethal form of disease of animals particularly that of cattle. It is one of the most communal economically significant infections of water buffalo and cattle in tropical areas of Asia, Africa and the Middle East. In Ethiopia also it is one of devastating diseases resulting in economic loss [17]. The disease alone does not result in high morbidity and mortality rate, but concurrent infection due to stressing factors like undernourishment, viral and bacterial attack of immune system, inclement weather, dehydration, nasopharyngeal settlement and shipment [18]. Tentative diagnosis can be made in many ways but, confirmation can be established by isolation of the causative organisms and their conventional and molecular characterization [19].
In Ethiopia the animal quarantine issue is underrated and is very poor due lack of policy regarding controlling system of the restriction of animal movement within the country and to the noubouring countries. Due to these cattle move with in the country without any restriction for marketing grazing and water. Likewise, cattle always come in the country from borders liker South Sudan where P. multocidasero type E2 and B2 causing HS are endemic. However, due to limited study, in Ethiopia particularly serotype E was not reported even though the suspection is very high. This is due to lack of detailed molecular based information on the prevailing serotypes at present. National Veterinary Institute (NVI) is producing only vaccine against Multocida type B2, and there is some complainton current available vaccine from some areas in Ethiopia, and this might be due to emerging of new strains which might be serotype E. Therefore, the molecular identification of the serotypes would improve our understanding of the epidemiology of strains involved to the disease as well as provide information on the most appropriate serotypes to be used for successful vaccination against bovine HS.
Thus, the general objective of the study is to determine the occurrences and molecularly characterize P. Multocida strains causing HS for the purpose of vaccine improvement.
The specific objectives are:
• Isolation and Identification of Strains P. Multocida from cattle in selected areas using conventional method.
• Molecular characterization of identified strains (Serotyping) for vaccine improvement.
Literature Review
Overview on is caused by P.Multocida
HS is a very acute, highly lethal form of disease of animals specifically buffalo, cattle, and bison. It is one of the most communal economically important infections of water buffalo and cattle in tropical areas of Asia, Africa and the Middle East, with erratic eruptions happening in southern Europe. The Disease is most devastating to smallholder farmers where husbandry and preventive practices are deprived and free-range management is common. Usual disease happens occasionally in sheep, pigs, and goats and has been conveyed in donkeys, camels, elephants, camels, yaks, and several species of deer and other wild ruminants [20].
Etiology
The Office International des-Epizoot (OIE) defined the conventional HS based on Carter and Heddleston classification system as the diseases caused by P. Multocida serotypes B:2 and E:2. Currently, serotype B:2 has been known in most areas where the disease is widespread. However, serotype E:2 has been found only in Africa [12].
Transmission
The common transmission route of HS from one animal to another is via inhalation or ingestion in the course of direct contact of adulterated feed and water. The causative bacteria are assumed to feast mostly in respiratory excretions, but they can also be establishing in other secretions and excretions, comprising feces and urine. Some animals remain carriers of the organisms in lymphatic tissues like in the tonsils, and shedding it periodically in nasal secretion during stress conditions [21].
Epidemiology
Three aspects affect the world wide circulation of HS: climatic circumstances, husbandry practices and the species of animal [22]. Later on disease eruption alive animals may come to be latent, asymptomatic carries of contagion. Herd immunity is high and so no new cases happen. Conversely, succeeding the contact of naïve animals, for instance, after animal travels, or the birth of new naïve animals, further disease outbreaks may occur. The infection was nearly non-existent where there was a prevalence of peaks. Intermittent sporadic eruptions occurred in areas with landscape, climate and animals that were between these dissipations [23].
Pathology and pathogenesis
On necropsy, the best evident gross lesion is subcutaneous oedema in the submandibular and brisket areas [24]. Petechial hemorrhages are originating subcutaneously and in the thoracic cavity. Furthermore, congestion and numerous degrees of consolidation of the lung might happen [24]. The death will happen contained by 24-36 hours, have only few petechial hemorrhages on the heart and wide spread blood accumulation of the lung, while in animals that die after 72 hours [24].
Geographic distribution
HS is an important infection of cattle and water buffalo in Asia, Africa and the Middle East. The primary incident is in Southeast Asia. The disease has likewise been transferred sporadically in Europe, where this disease is thought to occur frequently in southern Europe but has similarly continued observed rarely in further nations, counting those with humid weathers like Estonia, Latvia, Georgia and Ukraine. Numerous eruptions have remained conveyed as 2010 in Central Europe like Germany, Hungary and Serbia, signifying that the causal bacteria might have remained presented to this area after seemingly being lacking for eras [24]. HS-inducing serotypes of P. Multocida are not supposed to flow in North America, Australia or New Zealand. They remained similarly supposed to be lacking from South and Central America; nonetheless, this disease was conveyed to the World Organization for Animal Health (OIE) from Colombia in 2007, Venezuela in 2015 and Ecuador in 2018. There appears to be little or no published material around these eruptions [25,26].
Diagnosis
Clinical signs: HS diseased cattle develop peracute or an acute course of infection with signs of septicemia comprisingnasal discharge, malaise, recumbency, fever, dyspnoea, oedema and hemorrhages resulting in high mortality rates (OIE, 2012).
Postmortem lesions: HS is rapid in its course, therefore, the infected animals’ remains in good condition after death. Gross pathological lesions are mutable and might be understated and comprise circulatory and degenerative disorders. Liver looks yellowish-brown in pigment, hard and degenerated, with a clear lobular configuration. The tracheal mucosa is hyperemic, comprising plentiful frothy fluid, and the lungs are edematous and bloody. The spleen is enflamed, with smoothed boundaries and splenomegaly. The occurrence of coagulated blood in blood containers is due to dispersed intravascular coagulation [26].
Bacteriological method: Bacteriological medium like Blood agar, Nutrient agar, Casein-sucrose yeast (CSY) broth, Dextrose-starch agar, CSY agar CSY-blood agar, Enriched tryptophan agar and MacConkey agar (Oxoid, England), is used as selective medium because P. Multocidacannot grow on MacConkey. The colony character of P. Multocida like color, shape, size, consistency and a gram staining of culture media is considered. Conditions like growth reticence, motility; hemolytic property of growing bacteria was also seen [27]. The grams staining result under microscope reveals gram negative, small size rod shape of coccobacilli was being appreciated.
Serological technique: Serotyping using serological assays used comprise an agglutination test for somatic typing, rapid slide agglutination or indirect hemagglutination assays for capsular typing, agar gel immune diffusion for both capsular and somatic typing, and counter-immunoelectrophoresis for the rapid identification of capsular types B and E. Some sequesters can be hard to type with these traditional approaches [27].
Molecular identification: Molecular acid based diagnostic test has been establishing to be subtler and trustworthy than the conservative method of bacterial identification. The main benefit of nucleic acid based tests is that they decrease the time consumption and permit the discovery of the organism’s genome even if it is in tiny quantities, thus increasing the sensitivity and specificity of the test [28]. Polymerase chain reaction is one such test that can be used for the identification of organisms at any level, like strain, species, genus or all members of a domain, just by using explicit primer sequence [29].
Treatment
Antibiotics are the only effective drugs for the treatment of HS. However, they are only effective only if they are taken place immediately after the start of clinical signs. Monitoring the animals for febrile condition and treating them on the onset of the disease is advisable during the outbreak of the infection IV antimicrobials drugs like Oxytetracycline, pen-strep and various sulfonamides [30].
Control and prevention
Extended and unselective use of antibiotics against P. Multocida can lead to Multi Drug Resistant (MDR) [31]. Consequently, an effective control of disease can merely is attained by vaccination. Formalin-inactivated, alum-precipitated vaccine of P. Multocida currently the most broadly applied vaccine for preventing HS. Though, immunity persists for 4 to 5 months and protecting efficacy is only 60% [32].
Materials and Methods
Research area
The research was accompanied from December 2019 to August 2020 at two designated districts in Assosa Zone of Benishanigul Gumuz Regional State, namely Bambasi and Assosa. Assosa town is the clericalfocus of the Benishanigul Gumuz Regional State, and located 676 km West of Addis Ababa. Assosa zone is one of the 3 clerical zones of Benishanigul Gumuz Regional State. It is found in the Southwest part of the region. Assosa area is composed of 70 clerical farmworker associations and 4 Assosa town “kebeles” [33] which is located at 9.60° and 10.45° N and 34.20° and 34.58°E longitude. The elevation of Assosa arrays from 580 to over 1544 meter above sea level [34]. The entire zone of the district is 2317 Km2 of which area is categorized by low land flat agro-ecology rendering to National Meteorology Service Agency [34]. Bambasi district is situated in Benishanigul Regional State Southern west of the Assosa zone and 616 km west of Addis Ababa at 9.45-9.75°N and 34.35-34.88°E with a least and supremeelevation of 1350 m and 1770 m overhead sea level. Bambasi Woreda has the livestock populace 38964 cattle, 11,990 goats, 3452 sheep, 1995 donkeys, and 38442 poultry [35]. These regions were designated on the foundation of antiquity for occurrence of cattle HS, cattle drive to market, foraging land, drenching points and extent of cattle populace by the district office of livestock and fishery development.
Research design
The research design was a cross sectional study and it was intended for isolation of P. Multocida type B2 and E2. The study area was selected purposively based on epidemiological geographical location as Benishangul Gumuz regional state is the neighboring region to South Sudan where P. Multocida serotype E is endemic. Three visits were made to the study area purposively based on outbreak information. The choice of the animals was established on distinctive clinical signs of HS, and sampling of swabs and blood were completed from clinic and resident areas where eruption happened for isolation and identification purpose.
Research methods
Target population: The study population was local breed cattle infected with HS from the study area. The target populations were determined based on outbreak information of HS for the Isolation and identification of P. Multocida with the prominence of HS Causing Serotypes namely, B2 and E2 from indigenous cattle breeds.
Sampling technique: The technique used for sample collection from infected animals were purposive sampling and each animal was selected based on prominent specific clinical sings of HS from the study area where outbreak occurs and from those HS infected cattle brought to veterinary clinic in the study area. Two types of samples were collected namely, blood and swab samples. Swab was collected from nasal cavity of sick animals by introducing nasal swab about 5 cm deep in to the nasal cavity. Similarly, from those animals with high body temperature the blood was collected from jaguar vein after disinfecting the area with 70% alcohol.
Sample size: The sample size was determined based on the number of infected animals that were considered as an outbreak of HS in the study area. Thus, the purposive sampling method was used, therefore, the number of cattle sampled from Asosa, Bambasi and districts were 30 during the total three visits to the study area.
Data analysis method: For interpretation of the results, the whole data was entered into the Microsoft Excel sheet Data management and Analysist, and then it was analyzed using SPSS version 20. Regarding the molecular detection, the banding patterns of individuals’ strains were scored based on the presence or absence of the bands with the appropriate base pairs.
Isolation and identification of bacterial isolates
Cultural methods: All samples were plated onto nutrient agar (Oxoid, England) and incubated at 37°C overnight [36]. Single specific colony was taken and subculture on 10% sheep blood agar, then incubated at 37°C for 24 hours to observe the hemolytic effect of the organism. At the same time another single colony was taken to check the grams’ stains result under microscope. Sub culturing on selective medium called MacConkey agar (Oxoid, England) was done as P. Multocida do not grow on this media [37].
Biochemical methods: Succeeding biochemical tests were done for identification of P. Multocida. These tests include Oxidase, catalase, indole, H2S, motility and fermentation of glucose, mannitol, sucrose, Dextrose, maltose, lactose, using methods described by Cowan and Steel with modification of using CSY agar (Oxoid, England) [23,38,39]. The result found were recorded and compared for verification of the isolates to which species they were belong. The oxidase test was used to determine those organisms which possessed the cytochrome oxidase enzyme. The test was used as an aid for the differentiation of Neisseria, Moraxella, Campylobacter and Pasteurella species (oxidase positive).
A filter paper was soaked with the substrate Kovac’s oxidase reagent moisten the paper with a sterile distilled water. Then s single colony to be tested was placed on the filter paper. Finally Observation around inoculated area of paper for a color change to deep blue or purple within 10-30 second was detected. A negative reaction is indicated by absence of color. The Swab method was also done by sinking swab into reagent and then traces an isolated suspect colony and was detected for color change within 5-10 seconds and its result was similar with wet filter paper. Catalase test was done by putting a single colony of growth from culture onto a clean microscope slide. A few drips of H2O2 were added onto the smear. It was assorted with a tooth pick. A positive outcome was revealed the fast occurrence of O2 as showed by tangent foam formation [40].
Pathogenicity test: 24 hours’ growth from Tryphosa agar slant was suspended in 1 ml sterile physiological saline solution as labeled by Bain et al. [39]. The mean suspension density of each sample having 1 × 109 CFU/ml was evaluated at 540 nm. To evaluate the Pathogenicity of samples, rabbit was chosen and 0.5 ml growth suspension was injected by ear vein. Postmortem examination of the lifeless animal was done and re-isolation of the bacterium from the heart blood were cultured on Tryptose agar and incubated at 37°C for 24 hours.
Molecular identification: The pure growth of the bacteria was processed for molecular characterization through Polymerase Chain Reaction (PCR) using universal and capsular serotype specific primers of P. Multocida (Table 1) following [37].
| Type of primer | Direction | Name of primers | Sequences | Mole. weight |
|---|---|---|---|---|
| Universal primer | Forward primer | KMT1 KMT1T7 | 5’ATCCGCATTTACCCCAGTGG3’ | 460 bp |
| Reverse primer | KMT1SP6 | 5’-ATTC-3’ | ||
| Serotype | Forward primer | CAP E | 5’TCCGCAGAAAATTAATTGACTC3’ | 511 bp |
| E specific | Reverse primer | CAP E | 5’GCTTGCTGCTTGATTTTGTC-3’ | |
| Serotype | Forward primer | KTT72 | 5’AGGCTCGTTTGGATTATGAAG3’ | 620 bp |
| B specific | Reverse primer | KTSP61 | 5’ATTCCGCTAACACACTCTC3’ |
Table 1: Primers used for characterization of P. Multocida.
Results
Isolation and identification
Cultural characteristics of P. Multocida: Up on culturing of the total collected 30 blood and swab samples, it was revealed that only 4 samples out of 30 samples (13.3%) were found to show the cultural characteristics of p. multocida (Small, glistening, mucoid no hemolytic on blood agar and didn’t grow on MacConkey agar). All four isolates of Pasteurella are short coccobacilli with rounded ends, stained negative by Gram staining. Bipolar coloration of cells can be seen when smears of the studied culture are stained by Romanovsky-Giemsa method (Figures 1-4) [40].
Figure 1: Study area created by Arch GIS.
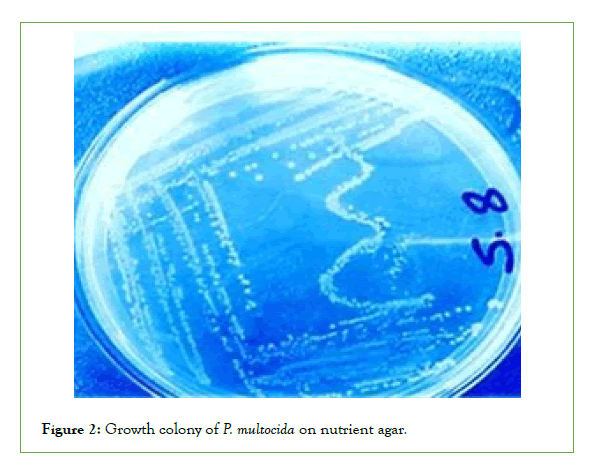
Figure 2: Growth colony of P. multocida on nutrient agar.

Figure 3: Grams’ stain isolated P. multocida.
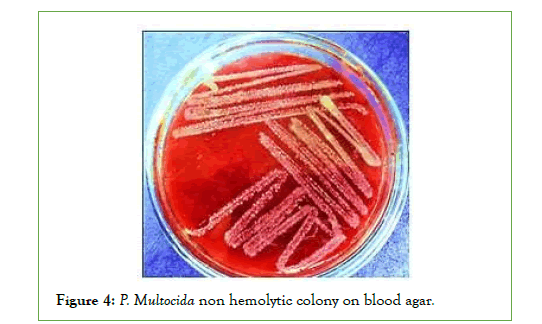
Figure 4: P. Multocida non hemolytic colony on blood agar.
Pathogenicity test activity of P. Multocida: With the aim of further confirmation like Pathogenicity study and acuity estimation, the 4 isolates were injected to Rabbit. All of the samples caused hemorrhages with in the heart and mortality in the animals. Replication of all of these tests on samples of died rabbit confirmed the isolates were Pasteurella (Figure 5).

Figure 5:Re-isolation of P. multocida from rabbit showing hemorrhage in the
heart.
Biochemical characteristics of P. Multocida: All the four isolates of biochemical characterization revealed that they belonged to Pasteurella genus. All strains isolated from Asosa and Bambasi districts were oxidase, Indole and catalase positive, did not procedure hydrogen sulfide, did not have hemolytic effect and were negative in Voges-Proskauer reaction. All isolates fermented glucose, Dextrose mannitol, sucrose, and were negative for maltose andlactose (Figures 6-10) [14].
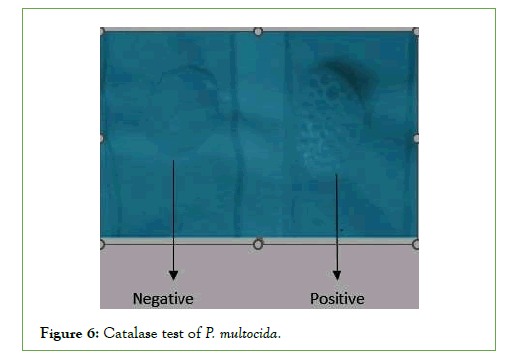
Figure 6: Catalase test of P. multocida.

Figure 7: Oxidase test of P. multocida.
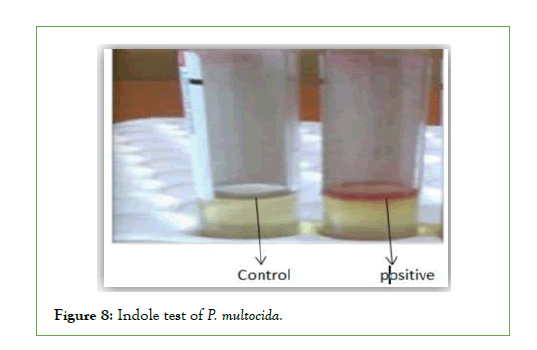
Figure 8: Indole test of P. multocida.

Figure 9: H2S and motility test of P. multocida.
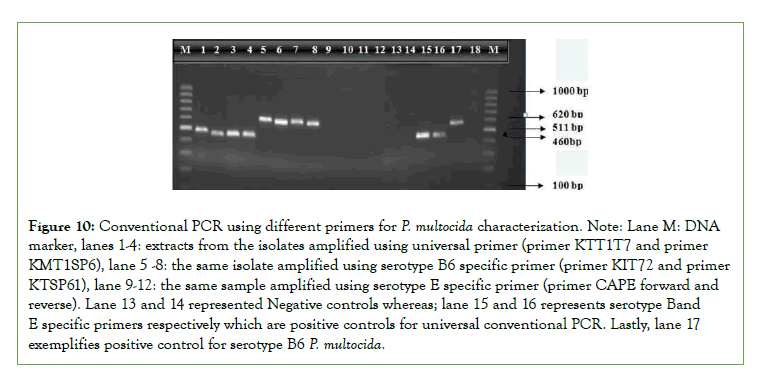
Figure 10: Conventional PCR using different primers for P. multocida characterization. Note: Lane M: DNA marker, lanes 1-4: extracts from the isolates amplified using universal primer (primer KTT1T7 and primer KMT1SP6), lane 5 -8: the same isolate amplified using serotype B6 specific primer (primer KIT72 and primer KTSP61), lane 9-12: the same sample amplified using serotype E specific primer (primer CAPE forward and reverse). Lane 13 and 14 represented Negative controls whereas; lane 15 and 16 represents serotype Band E specific primers respectively which are positive controls for universal conventional PCR. Lastly, lane 17 exemplifies positive control for serotype B6 P. multocida.
Discussion
Pasteurella multocida is an important pathogen causing a number of diseases in various domestic animals. The most important disease is HS resulting in heavy economic losses. This study was under taken at Benishangul regional state namely Asosa and Bambasi district aimed to isolate and identify P. Multocida serotypes resulting to HS by conventional and molecular methods. Up on culturing the organisms shows small, glistening, mucoid no hemolytic on blood agar and didn’t grow on MacConkey agar. Morphology by garm’s stain showed that it was gram negative coccobacilli under microscope. The biochemical Characteristics were oxidase, Indole and catalase positive, did not procedure hydrogen sulphide. All isolates fermented glucose, Dextrose mannitol, sucrose, and were negative for maltose and lactose. All these specific charactestics are similar to those described by Chesbrogh [41,42]. Similarly, Choudhury et al., [43] and Rahman [44] also stated the morphological features of p. multocida with grams’ negative and coccobacilli, with no haemolytic effect on blood agar which is similar to this study. Study by Cowan and Steel [38] also similar to this finding regarding the biochemical futures of P. Multocida like positive for Oxidase, catalase, Indole tests and negative for H2S. Shiva chandra [13] also reported similar biochemical characteristics for P. Multocida type B characteristics colonies of P. Multocida, as described by Choudhury et al., [43] and Yami et al., [17].With the aid of further confirmation like Pathogenicity study and acuity estimation on rabbit the selected organisms showed death and haemorrhage in the heart upon post mortem examination which is typical characteristics of pathogenic effect of P. Multocida as described in Bain et al. [39].
The overall isolation percentage of P. Multocida from the total number of samples examined was 3.39%. The various characteristics of P. multocida isolated during the present investigation are in conflict with the findings of Francis and Carter who reported the isolation of 17.5% P. Multocida from calves in Zambia [31]. This might be due to the small number of animals diagnosed due to the disease in this specific study. For confirmation of P. Multocida serotypes B:2 (6:B) and E:2 (6:E) as the principal causes of HS the PCR test was done based on Townsend et al. by using universal and specific primers [37].
Although serotype B: has been mainly reported in Asian countries and E:2 in African countries based on Townsend [37],both serotypes have been recovered from the disease in some African countries as Townsend [37]. Likely, in this study PCR remains only 100% specific for isolate of P. Multocida type B cultures with the predominant somatic antigen being either type 2 were identified by the amplification of a 620 bp fragment. However, the isolates in study area were not detected by conventional PCR using the amplification of an approximately 511 bp fragments with the CAPE forward and reverse serotype E primers which disagree with the study by Carteret., aland De Alwis [45], which says serotype E is the African serotype causing HS. Even though it is too early to exclude the effect of the emergence of new stains for ineffectiveness of the vaccines against Haemorrhagic Septicemia, lack of maintaining cold chain during transportation and storage of vaccines, inappropriate dose and route of administration and lack of strategic vaccination might be the cause of vaccine ineffectiveness [46,47]. However, still it is very important to cover a wide further epidemiological study area to conclude these suggestions.
Conclusion and Recommendations
In conclusion, HS is the most important disease of cattle in Africa caused by P. Multocida serotypes. The disease has been an extended standing delinquent with eruptions of the disease occurring mostly in the yearly basis. This is the preliminary study in Ethiopia, which comprises about the spread of both HS causing strains namely Serotype B and E at the same time in cattle in the field. Though, it was concluded that districts of study area were accepted as HS positive with serotype B specific and negative with serotype E specific. From the total sample collected from infected animals with HS 13% of samples were found to be positive for serotype B specific. However, no samples were found to be positive for serotype E specific. Isolates were highly pathogenic to rabbit. This study constitutes a crucial and initial step allowing in designing or improving the effectiveness and efficiency of the existing vaccine to support pasteurellosis disease controls in the country. Therefore, based on above conclusions the following recommendations were forwarded:
• The molecular investigation should be supported by the like DNA sequencing as to confirm P. Multocida strains that will ultimately helpful in designing an effective and efficient vaccine
• Too early to still recommend that P. Multocida serotype E is not circulating in Ethiopia, therefore, wide area coverage needed for further study in the area.
REFERENCES
- CSA R. The federal democratic republic of Ethiopia central statistical agency report on area and production of major. Statistical Bulletin. 2016.
- Kabeta T, Fikadu T, Zenebe T, Kebede G. Review on the pneumonic pasteurellosis of cattle. Acad J Anim Dis. 2015;4(3):177-184.
- Pasteur L. De l’attenuation du virus du cholera des poules. CR Acad Sci. 1880;91:673-80.
[Crossref], [Google Scholar]
- Pedersen KB, Barfod K. The aetiological significance of Bordetella bronchiseptica and Pasteurella multocida in atrophic rhinitis of swine. Nord Vet Med. 1981;33(12):513-522.
[Google Scholar], [Pub Med]
- Davies RL, Caffrey B, Watson PJ. Comparative analyses of Pasteurella multocida strains associated with the ovine respiratory and vaginal tracts. Vet Rec. 2003;152(1):7-10.
[Crossref], [Google Scholar], [Pub Med]
- Pedersen K, Dietz HH, Jørgensen JC, Christensen TK, Bregnballe T, Andersen TH. Pasteurella multocida from outbreaks of avian cholera in wild and captive birds in Denmark. J Wildl Dis. 2003;39(4):808-16.
[Crossref], [Google Scholar], [Pub Med]
- Marza AD, Abdullah FF, Ahmed IM, Chung EL, Ibrahim HH, Zamri-Saad M, et al. Involvement of nervous system in cattle and buffaloes due to Pasteurella multocida B: 2 infection: A review of clinicopathological and pathophysiological changes. J Adv Vet Anim Res. 2015;2(3):252-62.
[Crossref], [Google Scholar]
- Blanchong JA, Samuel MD, Goldberg DR, Shadduck DJ, Lehr MA. Persistence of Pasteurella multocida in wetlands following avian cholera outbreaks. J Wildl Dis. 2006;42(1):33-9.
[Crossref], [Google Scholar] [Pub Med]
- AHIS. Animal health information services: Deputy Commissioner (LH). Department of animal Health, Ministry of Agriculture, Govt. of India, New Delhi, India. 1997.
- Mustafa AA, Ghalib HW, Shigidi MT. Carrier rate of Pasteurella multocida in a cattle herd associated with an outbreak of haemorrhagic septicaemia in the Sudan. British Vet J. 1978;134(4):375-8.
[Crossref], [Google Scholar] [Pub Med]
- Singh VP, Kumar AA, Srivastava SK, Rathore BS. Significance of HS in Asia: India. International Workshop on Diagnosis and Control of HS. 1996:28-30.
- Ataei S, Burchmore R, Hodgson JC, Finucane A, Parton R, Coote JG. Identification of immunogenic proteins associated with protection against haemorrhagic septicaemia after vaccination of calves with a live-attenuated aroA derivative of Pasteurella multocida B: 2. Res Vet Sci. 2009;87(2):207-10.
[Crossref], [Google Scholar], [Pub Med]
- Biswas A, Shivachandra SB, Saxena MK, Kumar AA, Singh VP, Srivastava SK. Molecular variability among strains of Pasteurella multocida isolated from an outbreak of haemorrhagic septicaemia in India. Vet Res Commun. 2004;28(4):287-98.
[Crossref], [Google Scholar], [Pub Med]
- Quinn PJ, Carter ME, Markey B, Carter GR. Enterobacteriaceae. clinical veterinary Microbiology. 1994:109-35.
[Crossref], [Google Scholar]
- Jones RA, Mustafa AF, Christensen DA, McKinnon JJ. Effects of untreated and heat-treated canola presscake on milk yield and composition of dairy cows. Anim Feed Sci Technol. 2001;89(1-2):97-111.
- Catry B, Chiers K, Schwarz S, Kehrenberg C, Decostere A, De Kruif A. Fatal peritonitis caused by Pasteurella multocida capsular type F in calves. J Clin Microbiol. 2005;43(3):1480-3.
[Crossref] [Google Scholar] [Pub Med]
- Yami B, Kibeb L, Asmelash T. Isolation, identification and antimicrobial susceptibility of Pasteurella multocida from cattle with hemorrhagic septicemia in Asosa and Bambasi districts, Benishangul Gumuz Regional State, Ethiopia. Thesis research, AAU. 2017.
[Crossref]
- Hawari AD, Hassawi DS, Sweiss M. Isolation and Identification of Mannheimia haemolytica and pasteurella multocida in Sheep and Goats using Biochemical Tests and Heath protection agency, Indole test. National Standard Method BSO. 2008; 2:19.
[Crossref]
- Seleim RS. Major pathogenic components of Pasteurella multocida and Mannheimia (Pasteurella) haemolytica) isolated from animal origin. 2005.
- Bosch M, Garrido ME, Llagostera M, Pérez de Rozas AM, Badiola I, Barbé J. Characterization of the Pasteurella multocida hgbA gene encoding a hemoglobin-binding protein. Infect Immun. 2002;70(11):5955-5964.
[Crossref], [Google Scholar], [Pub Med]
- Benkirane A, De Alwis MC. Haemorrhagic septicaemia, its significance, prevention and control in Asia. Vet Med. 2002;47(8):234-240.
- Devendra C. Approaches to economic analyse s and implications for policy issues in South-east Asia: Results from three case studies in crop–animal systems. Economics Animal Health Prod. 2009:295.
- Quinn PJ, Markey BK, Leonard FC, Hartigan P, Fanning S, Fitzpatrick E. Veterinary microbiology and microbial disease. Can Vet J; 2002. [Crossref],
[Google Scholar], [Pub Med]
- De Alwis MC, Wijewardana TG, Gomis AI, Vipulasiri AA. Cultural, biochemical and Pathogenicity studies on P. Multocida isolated from carrier animals and from outbreaks of haemorrhagic septicaemia. Trop Anim Health Prod. 1990;22(3):185-194.
[Crossref], [Google Scholar]
- Tankaew P, Singh-La T, Titaram C, Punyapornwittaya V, Vongchan P, Sawada T, Sthitmatee N. Evaluation of an in-house indirect ELISA for detection of antibody against haemorrhagic septicemia in Asian elephants. J Microbiol Methods. 2017;134:30-34.
[Google Scholar], [Pub Med]
- OIE. Hemorrhagic septicemia; Office International des Epizooties, Paris, France. Terrestrial Manual, 2012:739–750.
- Holmes HT, Matsumoto M, Patton NM, Zehfus BR. Serologic methods for detection of Pasteurella multocida infections in nasal culture negative rabbits. Lab Anim. 1986;36(6):640-645.
[Google Scholar], [Pub Med]
- Innis I, Gelfand JJ, Sninsky, White TJ. PCR Protocols: A guide to methods and applications, edited by Michael A. Academic Press, 1990: 482: 39-95.
- Townsend KM, Boyce JD, Chung JY, Frost AJ, Adler B. Genetic organization of Pasteurella multocida cap loci and development of a multiplex capsular PCR typing system. J Clin Microbiol. 2001;39(3):924-929.
[Google Scholar], [Pub Med]
- OIE A. Manual of diagnostic tests and vaccines for terrestrial animals. Office international des epizooties, Paris, France. 2008:1092-1106.
- Varga Z, Sellyei B, Magyar T. Phenotypic and genotypic characterisation of Pasteurella multocida strains isolated from pigs in Hungary. Acta Vet Hung. 2007;55(4):425-34.
[Google Scholar], [Pub Med]
- Kehrenberg C, Schulze-Tanzil G, Martel JL, Chaslus-Dancla E, Schwarz S. Antimicrobial resistance in Pasteurella and Mannheimia: Epidemiology and genetic basis. Vet Res. 2001;32(3-4):323-339.
[Google Scholar], [Pub Med]
- ALFD. Assosa district office of livestock and fishery development. 2013.
- NMSA: National Meteorology Service Agency of Ethiopia. 2007.
- BLFD: Bambasi district office of livestock and fishery development. 2015.
- Kwage JK, Ekundayo SO, Chuku A, Yusuf AF, Mwankon ES, Boss SS, et al. Phenotypic and genotypic characterization of Pasteurella multocida isolated from dead poultry in Jos, Plateau State. Nigerian Vet J. 2013;34(2).
- Townsend KM, Frost AJ, Lee CW, Papadimitriou JM, Dawkins HJ. Development of PCR assays for species-and type-specific identification of Pasteurella multocida isolates. J Clin Microbiol. 1998;36(4):1096-1100.
[Google Scholar], [Pub Med]
- Cowan ST, Steel KJ. Manual for the identification of medical bacteria. 1965.
- Bain RVS, De Alwis MCL, Carter GR, Gupta BK. Hemorrhagic septicemia, Rome. FAO Animal Production and Health Paper .1982;33(8):256-260.
- Heath protection agency: Indole test. National Standard Method BSOP TP. 2010:2:19.
- Orynbayev M, Sultankulova K, Sansyzbay A, Rystayeva R, Shorayeva K, Namet A, et al. Biological characterization of Pasteurella multocida present in the Saiga population. BMC Microbiol. 2019;19(1):1.
[Google Scholar], [Pub Med]
- Cheesbrough M. District laboratory practice in tropical countries. Cambridge university press; 2005.
- Choudhury KA, Amin MM, Rahman A, Ali MR. Investigation of natural outbreak of fowl cholera. Bangladesh Vet J. 1985;19(1-4):49-56.
- Rahman MH, Shahiduzzaman ANM, Haque ME, Nazir NH, Rahman T. Random Amplified Polymorphic DNA (RAPD). Anal J Biological Sci. 2016;8:2.
- De Alwis MC. Haemorrhagic septicaemia. 1999.
- Carter GR, De Alwis MCL. Haemorrhagic septicaemia: Pasteurella and Pasteurellosis. Academic Press, London. 1989:131-160.
- De Alwis MC, Jayasekera MU, Balasunderam P. Pneumonic pasteurellosis in buffalo calves associated with Pasteurella multocida serotype 6: B. Ceylon Vet J. 1975;23:58-60.
Citation: Shuka J, Tesfay S, Ababae B, Abayineh T (2022) Isolation, Identification and Molecular Characterization of Pasteurella multocida from Cases of Cattle from Selected Areas. J Vaccines Vaccin. 13:478.
Copyright: © 2022 Shuka J, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

