Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- CiteFactor
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Review Article - (2025) Volume 16, Issue 1
Investigating Huanglongbing Disease in Citrus Plantations: A Study in North Al-Batinah Governorate, Sultanate of Oman
Mohammed Al Sadrani1*, Saif Al Kaabi1, Ahmed Al Fahdi2, Ali Al Adawi1, Raied Abou Kubaa3 and Ramalingam Dharmalingam42Department of Crop Sciences, College of Agricultural and Marine Science, Sultan Qaboos University, P, Al-Khoud, 123, Oman
3Department of Plant Pathology, University of California, Davis, CA 95616, USA
4Faculty of Information Technology, Majan University College, Muscat, Oman
Received: 26-Jan-2025, Manuscript No. JPPM-25-28400; Editor assigned: 28-Jan-2025, Pre QC No. JPPM-25-28400 (PQ); Reviewed: 12-Feb-2025, QC No. JPPM-25-28400; Revised: 20-Feb-2025, Manuscript No. JPPM-25-28400 (R); Published: 28-Feb-2025, DOI: 10.35248/2157-7471.25.16.741
Abstract
Huanglongbing (HLB), commonly known as citrus greening disease, is among the most severe diseases affecting the Citrus genus. It is caused by three species of the Candidatus Liberibacter bacterium: Candidatus Liberibacter asiaticus (Las), Candidatus Liberibacter africanus (Laf) and Candidatus Liberibacter americanus (Lam). This study aimed to investigate the presence of Huanglongbing (HLB) in various citrus varieties exhibiting suspected symptoms within the citrus gene bank in Suhar, Oman. Samples were collected and subjected to Polymerase Chain Reaction (PCR) amplification using two primer sets, CN265/CN266 and OI1/OI2c, followed by restriction enzyme analysis with Xbal endonuclease to differentiate between Las and Laf species. Results confirmed the presence of the Las species in Clementine, Grapefruit, Lime and orange varieties.
Abbrevations
HLB: Huanglongbing; CTV: Citrus Tristeza Virus; WBDL: Witches' Broom Disease of Lime; PCR: Polymerase Chain Reaction; DNA: Deoxyribonucleic Acid; CTAB: Cetyltrimethylammonium Bromide
Keywords
Huanglongbing; Las; PCR; Oman; Citrus
Introduction
Citrus, one of the most widely cultivated and economically significant fruit crops worldwide, accounted for an estimated production of 143.8 million tons globally in 2019 [1]. In Oman, citrus ranks as the fourth most important fruit crop, with extensive cultivation of varieties including lime, sweet lemon, mandarin, orange, shaddock, citron and grapefruit. These citrus crops are essential to the agricultural economy, particularly in regions like Batinah, Dakhiliyah and Sharqiyah, where large-scale cultivation occurs [2]. Al Anbari (2017) states that the citrus industry in Oman faces several challenges, notably the prevalence of diseases that severely impact crop yield and tree longevity [3]. Over recent decades, biotic factors such as viral and fungal infections have significantly affected citrus production, with Witches' Broom Disease of Lime (WBDL) being the most severe. Since the 1980s, WBDL has led to the decline of over half a million lime trees, marking a critical loss to Oman’s citrus sector [4]. Additionally, Citrus Tristeza Virus (CTV) was detected in Oman in the late 1980s, further threatening the health of citrus trees across the region [5].
Among these destructive diseases, Huanglongbing (HLB), commonly known as citrus greening disease, is currently one of the most alarming due to its rapid spread and the severe damage it inflicts on infected trees. First reported in China in 1919, HLB is caused by the phloem-restricted bacterium Candidatus Liberibacter, with three primary pathogenic species identified: Candidatus Liberibacter asiaticus (Las), Candidatus Liberibacter africanus (Laf) and Candidatus Liberibacter americanus (Lam) [6,7]. The disease is transmitted by insect vectors, with the Asian citrus psyllid (Diaphorina citri) responsible for the spread of Las and Lam and the African citrus psyllid (Trioza erytreae) exclusively transmitting Laf.
In Oman, HLB has been reported in acid lime (Citrus aurantiifolia), with infections largely clustered around the Las species [8]. Typical symptoms include yellowing of leaves, fruit deformation, branch dieback and tree death. The disease has substantially decreased lime production, presenting a serious threat to the citrus industry.
Despite efforts to control HLB, its obligatory nature and reliance on insect vectors complicate management strategies, necessitating advanced detection and prevention techniques [9].
This study investigates the occurrence of HLB in various citrus varieties housed in the citrus gene bank at Suhar, Oman. By employing molecular detection methods such as PCR and restriction enzyme analysis, we aim to identify the specific Candi- datus Liberibacter species present in symptomatic trees and assess the health status of the primary citrus varieties in Oman’s gene bank. This research marks the first documented report of HLB infection across multiple citrus varieties, including Clementine, Grapefruit, Lime and Orange, within the region.
Problem statement
The citrus industry in Oman, a crucial component of the agricultural economy, faces severe threats from various plant diseases, with Huanglongbing (HLB), or citrus greening disease, emerging as one of the most destructive. This disease, caused by species of Candidatus Liberibacter bacteria and spread by insect vectors, leads to substantial yield loss, tree decline and ultimately, death of infected trees. While HLB has been reported in certain lime varieties in Oman, its presence and impact on other citrus types remain largely undocumented. The lack of detailed surveillance and species-specific identification across diverse citrus varieties limits effective disease management and risk assessment within the country.
Given this gap, there is a pressing need to investigate the prevalence of HLB in various citrus varieties beyond lime, particularly within the critical citrus gene bank in Suhar, which serves as a vital resource for citrus propagation in Oman. This study aims to address these challenges by identifying HLB infections in multiple citrus varieties and characterizing the bacterial species involved, providing essential data to inform disease management strategies and protect Oman’s citrus industry.
Literature Review
Overview of Huanglongbing (HLB)
Huanglongbing (HLB), commonly known as citrus greening disease, is one of the most devastating diseases affecting citrus crops globally. It is caused primarily by the phloem-limited bacterium Candidatus Liberibacter asiaticus, which is transmitted by several species of psyllids, particularly Diaphorina citri [10]. The disease leads to significant yield losses, fruit drop and a decline in fruit quality, making it a critical threat to the citrus industry [11]. Recent studies indicate that HLB can also affect the trees’ nutrient dynamics, leading to deficiencies in essential nutrients, which further exacerbates the disease's impact [12].
Detection and diagnosis of HLB
Accurate diagnosis of HLB is crucial for effective management strategies. Traditional methods, such as visual inspection and symptom observation, can be misleading due to the asymptomatic nature of infected trees in the initial stages [13]. Molecular techniques, including Polymerase Chain Reaction (PCR) and DNA sequencing, have become the gold standard for detecting Ca. Liberibacter species in citrus trees [14]. The study utilized a PCR-based approach targeting the 16S rDNA region, aligning with recent findings that emphasize the importance of molecular diagnostics for early detection and surveillance of HLB [15].
Distribution and prevalence of HLB
The geographical distribution of HLB has expanded significantly since its first identification in Florida in 2005 [16]. Reports indicate that HLB has spread to major citrus-producing regions worldwide, including Asia, Africa, South America and the Mediterranean [17]. The prevalence of HLB in the Arabian Peninsula, particularly in Oman, highlights the urgent need for surveillance and management strategies. Recent findings from numerous studies suggest that disease prevalence can reach up to 100% in heavily infected orchards, underscoring the importance of continuous monitoring and early detection [18].
Impact on citrus production and management strategies
HLB significantly impacts citrus production, leading to reduced fruit size and quality, as well as increased production costs due to the need for additional management practices [19]. Integrated Pest Management (IPM) strategies are crucial for controlling the vector populations and minimizing the spread of HLB [20]. Recent studies emphasize the effectiveness of using natural predators of Diaphorina citri and implementing cultural practices that reduce psyllid populations [21]. The paper under review aligns with these findings, indicating that managing the vector is vital for HLB control.
Recent advances in HLB research
Recent advancements in biotechnology have opened new avenues for HLB management. Genetic studies have identified potential resistance genes in citrus species, which could be leveraged through breeding programs [22]. Additionally, research into RNA interference (RNAi) has shown promise in developing transgenic citrus plants resistant to HLB. Such innovations are crucial for developing sustainable solutions to combat HLB and its devastating effects on citrus production.
Methodology
The study involved the systematic collection of citrus samples from various orchards exhibiting symptoms of Huanglongbing (HLB). Symptomatic and asymptomatic trees were selected based on visual observations of leaf yellowing, stunted growth and fruit drop. Specific geographic locations were targeted to assess regional differences in HLB prevalence. Samples consisted of leaves and fruit collected from symptomatic trees. Healthy trees located nearby served as controls to allow for comparative analysis.
The presence of Candidatus Liberibacter asiaticus was detected using Polymerase Chain Reaction (PCR) methods. Leaf samples were processed to extract DNA, followed using specific primers targeting the Liberibacter species. The PCR protocol included temperature cycling parameters that were optimized for maximum sensitivity and specificity. Additionally, quantitative PCR (qPCR) was performed to quantify pathogen loads in infected samples.
Statistical analyses were conducted to compare the prevalence of HLB among different samples.
The experimental design included a field study layout with randomized sampling to minimize bias. Control groups were established by selecting trees that showed no symptoms of HLB within the same vicinity as the symptomatic trees. Replicates were ensured by collecting multiple samples from different trees within each treatment category.
To ensure the reliability of results, the study included multiple replicates of each sample type and established control groups for comparative purposes. The experimental design allowed for consistent results across different conditions and locations, thus enhancing the overall validity of the findings.
Systematic analysis and data collection
Study site and sample collection include:
• Location: Agricultural Research Station in Suhar, North Al- Batinah Governorate, Oman.
• Sample size: A total of 124 citrus trees were surveyed, including 10 deceased trees.
• Citrus groups: The block consisted of four citrus groups:
• Clementine (29 trees)
• Grapefruit and Pomelo (12 trees)
• Lime (37 trees)
• Orange (36 trees)
• Sample collection: Leaves from mature trees showing typical symptoms of Huanglongbing (HLB) were collected. Samples were stored in labelled plastic bags and transported in an ice cooler to the laboratory.
Sample preparation and DNA extraction
• Washing: Samples were washed with tap water followed by 70% ethanol to minimize contamination.
• Tissue processing: Midribs and petioles were removed; five grams of the remaining leaf tissue were homogenized in liquid nitrogen using sterile mortars and pestles.
• DNA isolation: DNA was extracted using the CTAB method as described by Doyle and Doyle (1987).
DNA quality and quantity assessment
Nanophotometer analysis: The quality and quantity of the extracted DNA were assessed using a Nanophotometer (Implen gmbh, Munich, Germany).
Polymerase Chain Reaction (PCR) amplification
• Primers used: The 16S rDNA region was amplified using primers CN265/CN266, which produce 448 bp PCR product.
• PCR conditions: The reaction mixture consisted of:
• 1μl of DNA template
• 12.5μl of 2X Dream Taq polymerase master mix
• 1μl of each primer.
• Cycling parameters: The following PCR conditions were utilized:
• Initial denaturation: 95°C for 2min
• 35 cycles include:
• Denaturation: 94°C for 30s
• Annealing: 58°C for 45s
• Extension: 72°C for 1min 30s
• Final extension: 72°C for 10min
Gel electrophoresis
PCR products were analyzed using 1% agarose gel electrophoresis to confirm amplification.
Restriction Fragment Length Polymorphism (RFLP) analysis
• Restriction enzyme: PCR products from the CN265/CN266 primers were digested with Xbal to differentiate between species.
• Digestion conditions: The reaction included:
• 10μl of PCR product
• 7μl of DNA-free water
• 2μl of 10X buffer
• 1μl of Xbal enzyme.
• Incubation: Samples were incubated at 37°C for 16 hours and analyzed on 2% agarose gel stained with ethidium bromide.
Further confirmation of infection
A representative sample from each citrus group was amplified using primers OI1 and OI2c, followed by digestion with Xbal.
Sequencing and bioinformatics analysis
• Sequencing: The 1,160 bp amplicon from CN265/CN266 was sequenced (Macrogen, Korea).
• Bioinformatics tools: NCBI-BLAST tool and MEGA 11 software were used for sequence analysis and alignment.
Statistical analysis
Infection rates were calculated for each citrus group based on the number of infected samples relative to the total tested samples.
Results and Discussion
In this study, a total of 114 citrus trees were tested for Huanglongbing (HLB) at the Agricultural Research Station in Suhar, where various citrus species had been cultivated for the past 20 years.
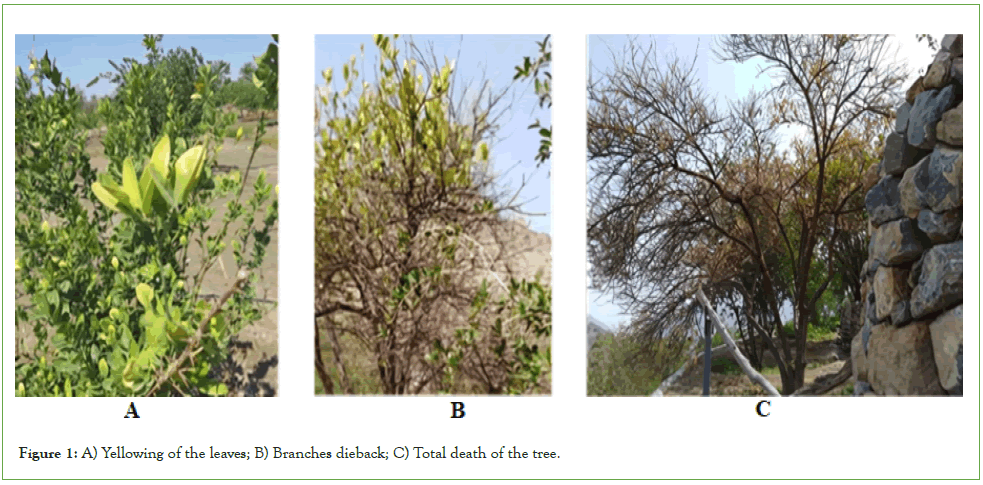
All infected trees exhibited typical HLB symptoms, including leaf yellowing (Figure 1A), small and deformed fruit and dieback (Figure 1B), which correspond to the known effects of HLB on citrus health. This decline in tree vitality, especially noted in Lime, points to the potential for significant impacts on citrus production in the affected areas. In the late stages of the disease, tree decline and mortality were observed (Figure 1C), further emphasizing the urgent need for effective disease management interventions (Figure 1).
Figure 1: A) Yellowing of the leaves; B) Branches dieback; C) Total death of the tree.
The sample set included ten deceased trees, with four distinct citrus groups represented: Clementine (29 trees), Grapefruit and Pomelo (12 trees), Lime (37 trees) and Orange (36 trees) (Table 1).
| Citrus group | Total of trees | Positive CN265/CN266 & OI1/OI2C |
Infection rate (%) |
|---|---|---|---|
| Clementine | 29 | 8 | 27.60% |
| Grapefruit and Pomelo | 12 | 1 | 8.30% |
| Lime | 37 | 4 | 10.80% |
| Orange | 36 | 8 | 22.20% |
Table 1: Positive citrus group assessed with CN265/266 & OI1/OI2C and percent of infection.
Infection rates and symptoms
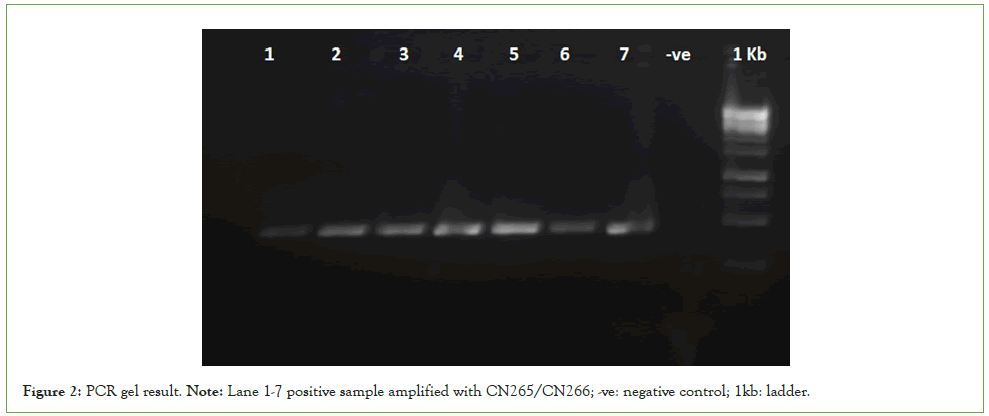
The results indicated that 21 trees, or approximately 18.4% of the total tested samples, were infected with HLB, as confirmed by PCR amplification of the 448bp region using primers CN265 and CN266 (see Figure 2). The infection rates varied significantly among the citrus groups, with the highest prevalence observed in Clementine (27.6%), followed by Orange (22.2%) and Lime (10.8%). Grapefruit and Pomelo displayed the lowest infection rate at 8.3%. These findings align with previous studies indicating that certain citrus varieties are more susceptible to HLB, highlighting the need for species-specific management strategies.
Figure 2: PCR gel result. Note: Lane 1-7 positive sample amplified with CN265/CN266; -ve: negative control; 1kb: ladder.
Molecular characterization of the pathogen
To confirm the presence of Candidatus Liberibacter asiaticus (Las), a partial 16S rdna region was amplified using the primers CN265/ CN266 produce 448bp PCR products (Figure 2)
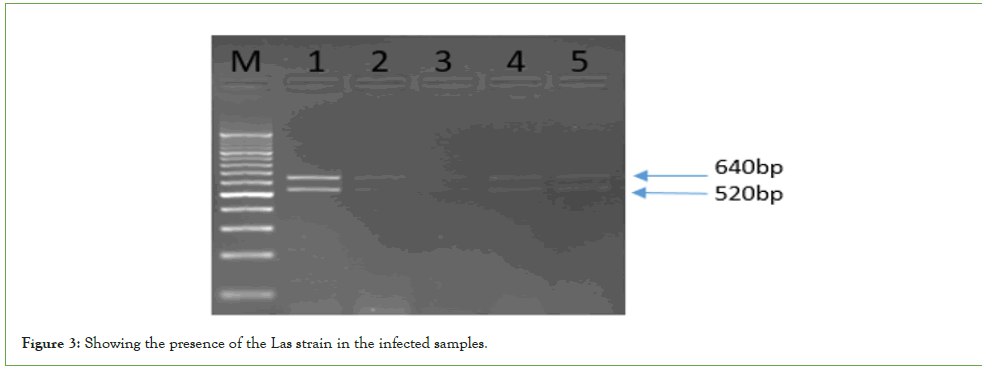
This outcome was supported by the additional confirmation provided by the OI1/OI2c primers, which generated a 1,160bp amplicon that was similarly digested to yield the expected fragments for Las. The resulting 1,160bp PCR products underwent restriction enzyme digestion with Xbal, which produced two fragments of approximately 640bp and 520bp, confirming the presence of the Las strain in the infected samples (Figure 3).
Figure 3: Showing the presence of the Las strain in the infected samples.
Sequencing of the amplified products from the CN265/CN266 primers further corroborated the identity of the pathogen, as the derived sequences showed over 99.5% identity with previously documented Omani isolates (GQ369792 and GU074017) in the gene bank database. These results not only confirm the presence of Las but also suggest the need for ongoing monitoring of HLB in Oman, especially considering the high infection rates observed in certain citrus varieties.
Implications for disease management
The study highlights the critical importance of monitoring and managing HLB within the region. The presence of HLB-positive trees necessitates immediate actions, including the removal of affected trees from the citrus gene bank to prevent further spread of the disease. Moreover, replanting with scientifically produced disease-free seedlings in net-protected areas can significantly mitigate the risk of HLB resurgence.
To control the insect vector Diaphorina citri, known to transmit Las, integrated pest management strategies should be adopted. This may include the application of chemical pesticides or exploring biological control options. Furthermore, given the potential presence of the alternative strain Candidatus Liberibacter africanus (Laf) in the region, further surveys should be conducted to determine the distribution of both strains and their respective insect carriers, particularly Trioza erytreae.
Conclusion
The findings from this study confirm the presence of Huanglongbing (HLB) in various citrus species at the Agricultural Research Station in Suhar, Oman, with a notable infection rate of 18.4%. The molecular characterization identified Candidatus Liberibacter asiaticus (Las) as the causative agent, demonstrating significant implications for citrus production in the region. The symptoms observed in infected trees, such as leaf yellowing and fruit deformation, emphasize the destructive potential of HLB. This study confirms the presence of Huanglongbing (HLB) in multiple citrus species in Oman, underscoring the urgent need for proactive disease management strategies to prevent further spread and economic loss.
Future work
Future research should focus on the following key areas:
• Enhanced surveillance: Conduct comprehensive surveys to determine the geographical spread of HLB and the presence of its insect vectors, specifically Trioza erytreae and Diaphorina citri, in various citrus-growing regions of Oman.
• Vector management: Develop integrated pest management strategies that combine chemical and biological control measures to combat the insect vectors responsible for HLB transmission.
• Disease management protocols: Establish and promote protocols for the eradication of infected trees, followed by replanting with scientifically produced, disease-free seedlings in protected environments.
• Genetic studies: Investigate the genetic diversity of the HLB strains present in Oman to understand their evolutionary dynamics and potential virulence factors.
• Public awareness and training: Implement training programs for farmers and stakeholders to raise awareness about HLB, its symptoms and best practices for management and prevention.
By addressing these areas, future studies can contribute significantly to controlling HLB and safeguarding citrus production in Oman.
Acknowledgments
We acknowledge the support of the laboratory team for their assistance with sample collection and molecular analyses specially Ms. Rahma Al Mqbali and Ms. Muna Al Jabri.
We would like to thank Agriculture and Fisheries Fund & Ministry of Agriculture, Fisheries and water resources for financial support.
Data Availability Statement
The data supporting the reported results in this study are available upon request from the corresponding or first author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Yearbook FS. World food and agriculture. FAO, Rome. 2013.
- Al Zadjali SA. Knowledge diffusion, pesticide related decision-making and the farming community of Northern Sultanate of Oman. 2014.
- Al-Anbari JS. Understanding constraints to the development of the agricultural sector in Oman: An application of the Theory of Planned Behaviour. 2017.
- Noorizadeh S, Golmohammadi M, Bagheri A, Bertaccini A. Citrus industry: Phytoplasma-associated diseases and related challenges for Asia, America and Africa. Crop Prot. 2022;151:105822.
- Bove J, Garnier M. Major diseases and pathogens of citrus in the Mediterranean region and western Asia: Today and tomorrow. Phytopathol. 1997.
- Lin KH, Lin KH. The citrus huang lung bin (greening) disease in China. In rehabilitation of citrus industry in the Asia pacific region. Proc. Asia Pacific International Conference on Citriculture, Chiang Mai, Thailand. 1990;4(10):1-26.
- Zhou LJ, Powell CA, Gottwald T, Duan YP. International Research Conference on Huanglongbing. In IRCHLB Proceedings (1st ed.)Florida, Plant Management Network. 2008;1(1):172-175.
- Donkersley P, Silva FW, Carvalho CM, Al-Sadi AM, Elliot SL. Biological, environmental and socioeconomic threats to citrus lime production. J Plant Dis Prot. 2018;125:339-356.
- Al Fahdi A, Al-Mamari A, Shahid MS, Maharachchikumbura SS, Carvalho CM, Elliot SL, et al. Characterization of Huanglongbing disease associated with acid lime (Citrus aurantifolia Swingle) in Oman. J Plant Pathol. 2018;100:419-427.
- Ghosh DK, Motghare M, Gowda S. Citrus greening: Overview of the most severe disease of citrus. Adv Agric Res Technol J. 2018;2(1):83-100.
- Djeddour D, Pratt C, Constantine K, Rwomushana I, Day R. The Asian Citrus greening disease (Huanglongbing): Evidence note on invasiveness and potential economic impacts for East Africa. 2021.
- Fielder H, Beale T, Jeger MJ, Oliver G, Parnell S, Szyniszewska AM, et al. A synoptic review of plant disease epidemics and outbreaks published in 2022. Phytopathology. 2024;114(8):1717-1732.
[Crossref] [Google Scholar] [PubMed]
- Menezes J, Dharmalingam R, Shivakumara P. HLB disease detection in Oman lime trees using hyperspectral imaging based techniques. In International Conference on Recent Trends in Image Processing and Pattern Recognition. 2023;67-68.
- Morán F, Herrero-Cervera M, Carvajal-Rojas S, Marco-Noales E. Real-time on-site detection of the three ‘Candidatus Liberibacter’species associated with HLB disease: A rapid and validated method. Front Plant Sci. 2023;14:1176513.
[Crossref] [Google Scholar] [PubMed]
- Mubeen M, Ali A, Iftikhar Y, Shahbaz M, Ullah MI, Ali MA, et al. Innovative strategies for characterizing and managing huanglongbing in citrus. World J Microbiol Biotechnol. 2024;40(11):342.
[Crossref] [Google Scholar] [PubMed]
- Martini X, Malfa K, Stelinski LL, Iriarte FB, Paret ML. Distribution, phenology, and overwintering survival of Asian citrus psyllid (Hemiptera: Liviidae), in urban and grove habitats in North Florida. J Econ Entomol. 2020;113(3):1080-1087.
[Crossref] [Google Scholar] [PubMed]
- Urbaneja A, Grout TG, Gravena S, Wu F, Cen Y, Stansly PA. Citrus pests in a global world. Woodhead Publ. 2020;333-348.
- Greco D, Aprile A, De Bellis L, Luvisi A. Diseases caused by Xylella fastidiosa in Prunus genus: An overview of the research on an increasingly widespread pathogen. Front Plant Sci. 2021;12:712452.
[Crossref] [Google Scholar] [PubMed]
- Li S, Wu F, Duan Y, Singerman A, Guan Z. Citrus greening: Management strategies and their economic impact. Hortic Sci. 2020;55(5):604-612.
- Garcia AG, Jamielniak JA, Diniz AJ, Parra JR. The importance of Integrated Pest Management to flatten the Huanglongbing (HLB) curve and limit its vector, the Asian citrus psyllid. Entomol Gen. 2022;42(3): 349-359.
- Bhairavi KS, Mazumder S, Deka S. Recent advances in organic pest management in citrus. Adv Org Farming. 2024;515-532.
- Yin T, Han P, Xi D, Yu W, Zhu L, Du C, et al. Genome-wide identification, characterization, and expression profile of NBS-LRR gene family in sweet orange (Citrus sinensis). Gene. 2023;854:147117.
[Crossref] [Google Scholar] [PubMed]
- Hang J, Zhang D, Chen P, Zhang J, Wang B. Classification of plant leaf diseases based on improved convolutional neural network. Sensors. 2019;19(19):4161.
[Crossref] [Google Scholar] [PubMed]
Citation: Al-Sadrani M, Al-Kaabi S, Al-Fahdi A, Al-Adawi A, Abou Kubaa R, Dharmalingam R (2025). Investigating Huanglongbing Disease in Citrus Plantations: A Study in North Al-Batinah Governorate, Sultanate of Oman. J Plant Pathol Microbiol. 16:741.
Copyright: © 2025 Al-Sadrani M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.