Indexed In
- Academic Journals Database
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- China National Knowledge Infrastructure (CNKI)
- Scimago
- Ulrich's Periodicals Directory
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- MIAR
- University Grants Commission
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2014) Volume 5, Issue 4
Immunogenicity of Leptospira interrogans Outer Membrane Vesicles in a Hamster Model
Abstract
Animal Leptospirosis, caused by Leptospira spp., is one of the most common zoonotic diseases in the US and throughout the world. Commercially available bacterins are only partially efficacious in that they protect against death, but not against disease. Protective immunity to Leptospira spp. require antibodies specific to outer surface proteins and/or adhesins of leptospires. Spirochetes produce membrane blebs or vesicles (OMVs) and OMVs have been shown to be good immunogens. In this study, we characterized leptospiral OMV components by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of and identified that the majority (58.1%) of proteins in the vesicles were cytoplasmic proteins (294 of 506), while 5 were extracellular proteins (0.99%), 11 were outer membrane proteins (2.17%), 14 were periplasmic proteins (2.77%), 48 were cytoplasmic/inner membrane proteins (9.49%) and 134 were unknown or having multiple locations (26.48%). Transmission electron microscopy (TEM) imaging showed OMVs are spherical bodies with a diameter of 50-200 nm. Vesicles were used to vaccinate hamsters. The results indicated that immunization with Leptospira OMVs induced significant protection against lethal challenge revealed by an enhanced humoral immune response, high survival rate and significantly reduced bacterial burden, all of which were reflected in decreased pulmonary, hepatic and renal lesions (p<0.05). To the best of our knowledge, this is the first report showing that OMVs could be used as a novel vaccine formulation to protect hamsters against lethal challenge.
Keywords: Outer membrane vesicles, Immunogenicity, Vaccine, Proteomics, Leptospira interrogans
Introduction
Pathogenic Leptospira spp. cause leptospirosis a zoonotic disease that is prevalent in people, dogs, pigs, horses, cattle, sheep, goats and wild animals [1,2]. The disease occurs widely in developing countries, such as Thailand, Brazil and India, and is reemerging in developed countries [3-7]. Although the incidence of human leptospirosis in the United States is relatively low, disease incidence in domestic animals has increased in recent years [3]. Leptospiral infection can range in severity from an inapparent infection to death from pulmonary hemorrhage, renal disease or hepatic failure [1,2,8,9]. Leptospiral infection can also cause uveitis and is considered the leading cause of blindness in horses [10,11]. Infection is acquired primarily through contact of mucous membranes or broken skin with water, moist soil or vegetation contaminated with the urine of infected animals. In 2013, leptospirosis became a reportable disease in the United States [12].
Although leptospirosis can be cured with antibiotics, treatment with antibiotics in the late stage of the disease may result in permanent kidney or lung disease that may impair the patient’s health and wellbeing. The excessive use of antibiotics in livestock raises food safety issues. Therefore prophylactic vaccination, along with the application of preventive strategies, seems to be the most convenient and effective way to control the disease. Newer vaccines against canine leptospirosis include multiple serovars of inactivated pathogenic leptospires induce protective immune responses [13,14]. However, there is still room for improvement in the durability of protective immunity and cross serovar protection [4,13,15,16]. A vaccine consisting of multiple letpospiral OMPs [17-22] has been shown to elicit significant immunological responses and provides at least some protection.
During the past decade, outer membrane vesicles (OMVs) have become increasingly attractive as an alternate vaccine preparation for infectious diseases, especially those caused by Gram negative bacteria [23,24]. OMVs are enclosed lipid bilayer structures that are usually enrich in outer membrane elements necessary for bacterial virulence and pathogenesis, such as outer membrane proteins, virulence factors, and enzymes [25,26]. These virulence factors could act as antigens and contribute to immune responses. In vitro and in vivo studies revealed that OMVs are able to bind to host cells and mediate the initial stages of infection by delivering virulence factors or other immunomodulatory molecules into the cells, often through internalization of entire OMVs. Internalizaiton can trigger inflammatory cascades and immune responses against the pathogens [27]. Based on their unique characteristic and the presence of virulence factors, OMVs are promising novel vaccine candidates that might provide stronger and more efficacious immune responses. An example of a highly successful bacterial membrane vesicle-based vaccine that used to prevent N. meningitidis serogroup B infection [23]. Several different formulations of this vaccine have been developed in order to target different strains and antigens existing in specific geographic regions. All formulations are able to activate mucosal and systemic bactericidal antibodies. The concept of developing an OMV based vaccine has also been established for other bacterial diseases: Bordetella pertussis [28], Borrelia burgdorferi [24], Burkholderia pseudomallei [29], Francisella tularensis [30] and Vibrio cholera [31]. Humoral and/or cellular immunity towards the vesicles and sufficient protection were obtained in immunized animals, suggesting the potential use of OMVs either as antigens and/or vaccine delivery vehicles.
In this study, we isolated OMVs isolated from pathogenic L. interrogans serovar Pomona and evaluated their efficacy as an acellular vaccine against leptospirosis in a hamster model. Leptospira OMVs contained multiple well-established outer membrane antigens and immunization with OMVs clearly induced the significant protection upon the challenge. Our results suggest that Leptospira OMVs have promise as an effective vaccine that could be used for leptospirosis prevention and control.
Materials and Methods
Bacterial strain
L. interrogans serovar Pomona (NVSL11000) used for challenge was obtained from the National Veterinary Service Laboratory (NVSL), Ames, Iowa. Leptospires were maintained in EMJH medium at 30°C. The low passage cultures were isolated by experimentally infecting hamsters with a sublethal dose of L. interrogans serovar Pomona as previously described [32].
Animals
Female Golden Syrian hamsters (Harlan Sprague Dawley) 3-4 weeks old were used in the study. The animals had ad libitum access to a commercial pelleted ration and drinking water. Experiments were conducted according to the protocol approved by IACUC (Institutional Animal Care and Use Committee) at Cornell University.
Production of outer membrane vesicles
OMVs were purified following the method of Nally et al. [33] with slight modifications. Briefly, Leptospira cultures, grown in EMJH media at 30°C until reaching mid to late log phase, were harvested by centrifugation at 12,000xg at room temperature for 40 min. Pellets were washed twice by re-suspension in phosphate-buffered saline (PBS), pH 7.4 and centrifuged at 13,000xg for 20 min. The pellets were then re-suspended in ice-cold 0.1 M citrate buffer, pH 3.0 and shaken vigorously for 3 h with vortex for 2 min at 20-min intervals. Samples were then centrifuged for 10 min at 1,000xg and pellets were re-suspended in a final volume of 4-6 ml 0.1 M citrate buffer, pH 3.0 depending on weight of the pellets. Samples were passed through a French press at 12,000 psi and monitored by dark-field microscopy. Lysed samples were centrifuged for 5 min at 200xg. The supernatant was added to an 11 ml 20–60% continuous sucrose gradient and centrifuged for 16 h at 77,000xg, 4°C. The upper OMV band was harvested by piercing the tube with an 18-gauge needle on a 10 ml syringe. OMV was washed using PBS, pH 7.4 and then pelleted by centrifugation at 103,000xg, 4°C, for 4 h. The supernatant was removed and the pellet was re-suspended in PBS, pH 7.4. The protein concentration of purified OMVs was then measured using the Bradford method.
Transmission electron microscopy
Images of OMVs were taken with the FEI Tecnai™ G2 Spirit Twin transmission electron microscope at Cornell Center for Materials Research (CCMR). A droplet of OMVs suspended in PBS was spotted on a grid coated with a carbon-reinforced fomvar film. The excess fluid was then removed with absorbent filter paper and the grids were stained with 2% (w/v) phosphotungstic acid pH 5.2 (with KOH). Vesicles were visualized under the TEM.
Liquid chromatography mass spectrometry analysis of OMVs
Ten micrograms of OMVs isolated from the bacteria were separated on 12% SDS-PAGE. Coomassie blue stained gel bands were manually excised into small pieces and transferred to microcentrifuge tubes. Gel pieces were washed with 100 µl milli-Q H2O for 5 min, 100 µl 100 mM ammonium bicarbonate in 50% acetonitrile for 10 min and 50 µl acetonitrile for additional 5 min. After washing, gel pieces were dried in a vacuum centrifuge or ventilated fume hood. Gel samples were next incubated in 20 µl of 10 mM DTT in 100 mM ammonium bicarbonate for 45 min at 56°C, followed by incubation in the dark in 20 µl of 55 mM iodoacetamide in 100 mM ammonium bicarbonate for 30-60 min. Wash steps were repeated and in-gel proteins were digested with trypsin in 50 mM ammonium bicarbonate at 30°C overnight. Extraction of peptides was performed by adding formic acid to a final concentration of 1%. Supernatant was transferred to a new tube and 30 µl of 5% formic acid in 50% acetonitrile was added to the samples, incubated for 20 min, and the supernatant was saved. This step was repeated once, followed by incubating in 5% formic acid in 90% acetonitrile. Total supernatant extracts were dried using a SpeedVac (Thermo Savant, Holbrook, NY). Peptides were reconstituted, loaded and separated using an UltiMate 3000 RSLCnano (Dionex, Sunnyvale, CA) on C18 columns coupled with a LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, Waltham, MA) equipped with a nano ion source. Identification of peptides was performed by raw MS/MS spectra search with licensed Mascot Deamon version 2.2.04 (Matrix Science, Boston, MA) containing complete Leptospira protein reference sequences of 2 L. interrogans serovars, 2 strains of L. borgpetersenii serovar Hardjo-bovis, and 2 strains of L. biflexa serovar Patoc downloaded from the NCBI reference sequence database. Significant score for the peptides was defined by a Mascot probability analysis with randomized peptide identification probability <0.01.
PSORTb version 3.0 [34] with Gram negative option was utilized for the prediction of subcellular localization of each identified protein in the samples. The final prediction site was determined based on the confidence values for each of the localization sites (a score of 7.5 or greater) and returned as one of five subcellular locations: extracellular, outer membrane, periplasm, inner/cytoplasmic membrane or cytoplasm. If two sites had high scores, a flag of "This protein may have multiple localization sites" was also returned in the Final Prediction field. LipoP 1.0 [35-37], a prediction algorithm for lipoprotein signal peptides, was applied to each identified protein in order to confirm the presence of a signal peptide in the sequences.
Immunization
Golden Syrian hamsters (3-4-weeks-old) were divided into 2 groups consisting of 8 animals each. One was a control group [PBS administered with 2% Alhydrogel (Invivogen, CA) at a final concentration of 1% (v/v)], while the second group was OMVs-vaccinated. Prior to immunization, animals were anesthetized by intraperitoneal injection of 0.1 ml of ketamine/xylazine (ketamine, 100 mg/ml; xylazine, 100 mg/ml) per 135 g hamster weight. The animals were immunized 3 times at a 3-week interval subcutaneously with 50 µg of total proteins in 150 µl volume. Animals were bled through the saphenous vein on day 0 (pre-vaccination, PV), 21 (before 1st boost), 42 (before 2nd boost), 63 (before challenge, BC) and 84 (after challenge, AC). Sera were collected and kept at -80°C until use.
Challenge
The Leptospira challenge was performed as previously indicated [32]. Three weeks after last booster, each animal was injected intraperitoneally with 1.5 ml PBS containing 2.5×102 (2.5× modified LD50 [MLD50]) of a single passage of L. interrogans serovar Pomona. The hamsters were observed twice daily for 3 weeks for clinical signs and mortality. If the hamsters showed severe clinical signs (moribund) they were euthanized for blood and tissue collection and counted as dead. Hamsters that survived the challenge were sacrificed at the end of the observation period and bled by cardiac puncture. Sections of kidney, liver and lung were collected for histopathology. For Leptospira culture, kidney, liver and urinary bladder were aseptically isolated.
Antibody response determination by ELISA
The antibody response against various vesicle proteins was analyzed by enzyme-linked immunosorbent assay (ELISA) [38]. Briefly, wells of polystyrene plates (Nunc, Denmark) were coated overnight with 1 µg of OMV antigens in 100 µl coating buffer (carbonate-bicarbonate buffer, 0.1 M, pH 9.6) per well at 4°C. After washing 3 times with PBS, plates were blocked with 200 µl blocking buffer (5% non-fat dry milk in PBS) and incubated for 1 hour at 37°C in a humid chamber. Then, 100 µl of hamster serum, diluted 1: 500 in blocking buffer, were added and incubated for 2 hours at room temperature. This was followed by washing and incubating with 100 µl of peroxidase labeled goat anti-hamster IgG antibody at 1: 8,000 dilution (KPL, Gaithersburg, MD). Finally, plates were washed and 100 µl of tetramethyl benzidine (TMB) substrate solution (KPL) were added to each well. After color development, the absorbance was read in microplate reader at 630 nm (Bio-Tek, Winooski, VT).
Quantification of Leptospira load in tissues using real-time PCR
Liver, kidney and urinary bladder were aseptically removed from each infected animal that died after challenge and from the animals that survived through the end of the experiment. The same organs from uninoculated hamsters were also collected and used as controls. All tissue samples were kept at -80°C until used. To determine bacterial load, total genomic DNA from the frozen tissues was isolated using the DNeasy Blood & Tissue Kit (QIAGEN, Valencia, CA) following the manufacturer’s protocol with a final 200 µl elution. Quantitative real-time PCR was performed using Taqman assay with primers-probe, targeting conserved region of ligA and ligB, designated as ligCon, of Leptospira serovars [39]. LigCon primers-probe were designed using PrimExpress software (Intergrated DNA Technology, Coralville, IA). Sequences of forward primer, reverse primer, and probe were: ligConF-5'AGCCATCGGTATCTTTTCGG3', ligConR-5'TGCACGAATATGAGCTGTTCC3', and ligConP-5'CCCTGAGGCCAACCCTGAATCAT3'. The 5' end and 3' end of probes were labeled with 6-carboxyfluorescein (FAM) and BHQ-1 (Integrated DNA Technology) respectively. All real-time PCR were set up in a 20 µl reaction volume consisting of the following reagents: 1×TaqMan® Gene Expression Master Mix (Applied Biosystems, Foster City, CA), 400 nM of each primer, and 200 nM probe with 5 µl of gDNA sample for each reaction. Amplification and fluorescence detection were performed in 7500 Fast Real-Time PCR (Applied Biosystems) platform operating on standard conditions (50°C for 2 min and 95°C for 10 min followed by 40 cycles of 95°C for 15 s, 60°C for 1 min). All reactions were conducted in duplicate with negative and positive controls for each run. The data was analyzed using 7500 Fast Real-Time PCR System Software (Applied Biosystems).
The standard curve was prepared by serial dilution of 107–100 plasmid copies and subjected to real-time PCR along with the samples for each run. Standard curves for quantification of leptospires were established by plotting the copy numbers of recombinant plasmid – pGEX4T2 containing conserved region of ligA and ligB (pligCon) against the threshold cycle (CT). Number of leptospires in the samples was then quantified by calculating CT values against copy numbers in the standard curve (there are two copies of ligCon region per Leptospira genome, from ligA and ligB; therefore the obtained copy numbers were divided by 2 to get the absolute number) and reported as the number of leptospires/milligram tissue.
Histopathological study
Hamster tissues were collected and fixed by immersion in 10% neutral buffered formalin. The fixed tissues were sectioned at 5 µm, stained with hematoxylin and eosin, and examined by light microscopy. The severity of Leptospira induced lesions was graded by a board certified veterinary pathologist (S.P. McDonough) who was blinded to treatment group. Tubulointerstitial nephritis was assessed as 0=normal, 1=mild, 2=moderate, and 3=severe, using the same criteria as previously described [34]. Hepatic lesions were graded based on the number of inflammatory foci present in 10 random 10×fields: 0=normal, 1=1–3, 2=4–7 and 3=>7. Severity of pulmonary hemorrhage was graded as 0=normal, 1=focal, 2=multifocal, and 3=locally extensive.
Culture
To determine conclusively the presence of Leptospira, liver, kidney, and urinary bladder of infected animals were also submitted for culture in EMJH medium and maintained at 30°C for 4 weeks. Growth of leptospires was monitored using dark-field microscopy. The culture samples showing visible growth of leptospires were considered positive and those without visible growth were reported as negative.
Statistical analysis
To compare differences in antibody response, leptospiral load and degree of tissue inflammation in vital organs, the two-tailed t-Test included in Excel software was used to perform statistical analysis. A p value of <0.05 was considered statistically significant.
Results
Visualization and proteomic analysis of Leptospira interrogan vesicles
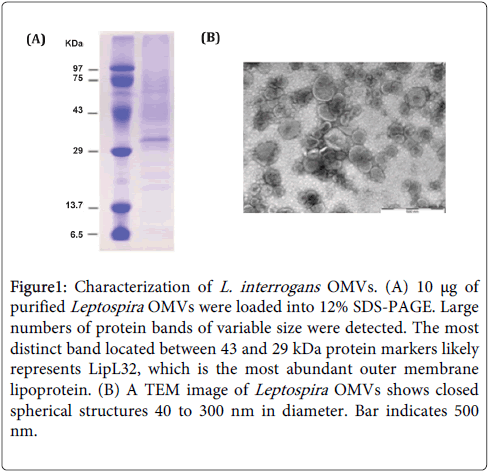
By incubation of whole leptospire cells in low pH citrate buffer, passing through a French press to lyse the cells, and employing ultracentrifugation over a continuous sucrose gradient, fragmented outer lipid bilayer membranes were enclosed and formed Leptospira OMVs. The yield of OMVs is ~ 0.3 mg per liter of leptospiral culture reproducibly. 1-D SDS-PAGE analysis and Coomassie Blue staining of total protein bands revealed a large number of proteins present in the vesicles with a wide molecular mass range: ~ 6 to >100 kDa (Figure 1A). TEM examination with negative staining confirmed that Leptospira OMVs purified by this method are double membrane spherical structures and vary in size (40-300 nm) (Figure 1B). To further determine protein composition in the OMVs, nanoLC-MS/MS analysis of total vesicular proteins separated by 1-D electrophoresis was done. A total of 506 L. interrogans proteins with the significant ion score equal to or above the threshold (p<0.005) were identified by performing spectra search against customized Mascot server containing complete Leptospira NCBI protein RefSeq database, listed in Supplement data (Table 1). PSORTb analysis of all matched proteins for their subcellular location indicated that the majority (58.1%) of proteins in Leptospira vesicles were characterized as cytoplasmic proteins (294 of 506), whereas 5 were described as extracellular proteins (0.99%), 11 as outer membrane proteins (2.17%), 14 as periplasmic proteins (2.77%), 48 as cytoplasmic/inner membrane proteins (9.49%) and 134 as unknown or having multiple locations (26.48%). Some the Leptospira lipoproteins, for example Lig proteins, have a unique signal peptide sequence that is different from the signal peptide found in other bacterial species and may not be recognized by PSORTb. Therefore, LipoP 1.0 algorithm for prediction of lipoprotein signal peptide and discrimination between lipoprotein signal peptides, other signal peptides and n-terminal membrane helices in Gram-negative bacteria was used to confirm the presence of signal peptides in all identified proteins. LipoP determined 54 (10.67%) proteins as lipoproteins as they carried signal peptide type II, 64 proteins (12.65%) with signal peptide type I and the rest,388 (76.68%), were predicted to have no signal peptide.
Figure 1: Characterization of L. interrogans OMVs. (A) 10 μg of purified Leptospira OMVs were loaded into 12% SDS-PAGE. Large numbers of protein bands of variable size were detected. The most distinct band located between 43 and 29 kDa protein markers likely represents LipL32, which is the most abundant outer membrane lipoprotein. (B) A TEM image of Leptospira OMVs shows closed spherical structures 40 to 300 nm in diameter. Bar indicates 500 nm.
| Accession No. | Protein Description | Protein | Matched | Sequence | MW (kDa) | pI | PSORTba |
|---|---|---|---|---|---|---|---|
| Score | Peptides | Coverage (%) | |||||
| NP_712836.1 | Molecular chaperone GroEL | 6528 | 219 | 73.3 | 57.99 | 5.29 | CY |
| WP_000372289.1 | Fe-S-cluster-containing hydrogenase | 6392 | 180 | 44.7 | 113.74 | 8.46 | CY |
| WP_000736495.1 | Membrane protein LipL32 | 6261 | 227 | 63.2 | 29.68 | 6.34 | UN** |
| NP_710192.1 | Outer membrane lipoprotein LipL21 | 5590 | 221 | 57.5 | 19.93 | 7.53 | PP** |
| YP_002879.1 | LipL41 | 5583 | 170 | 62.8 | 39.09 | 7.52 | UN** |
| NP_712320.1 | Malate dehydrogenase | 4613 | 171 | 71.8 | 35.12 | 6.92 | UN |
| WP_000959947.1 | Endonuclease | 3871 | 115 | 65.4 | 35.77 | 9.16 | UN |
| NP_710918.1 | Elongation factor Tu | 3711 | 139 | 65.1 | 43.66 | 5.74 | CY |
| WP_000695325.1 | F0F1 ATP synthase subunit alpha | 3643 | 133 | 46.3 | 55.12 | 5.68 | CY |
| NP_712198.1 | Endoflagellar filament core protein | 3630 | 107 | 57.7 | 31.29 | 6.54 | PP |
| WP_001079039.1 | Hypothetical protein | 3587 | 136 | 66.5 | 44.79 | 7.55 | UN** |
| YP_000182.1 | Hypothetical protein LIC10191 | 3188 | 120 | 75.4 | 20.58 | 8.56 | UN** |
| YP_001212.1 | F0F1 ATP synthase subunit beta | 3052 | 106 | 72.6 | 50.59 | 5.56 | CY |
| WP_002103311.1 | Acetyl-CoA C-acetyltransferase | 2871 | 133 | 74.8 | 47.47 | 6.14 | CY |
| WP_000645001.1 | Cysteine synthase | 2833 | 104 | 59.5 | 33.24 | 7.82 | CY |
| NP_712064.1 | Hypothetical protein LA1883 | 2578 | 90 | 39.4 | 58.42 | 8.86 | CY** |
| YP_003074.1 | Hypothetical protein LIC13166 | 2570 | 73 | 35.9 | 36.3 | 8.5 | UN/MS* |
| WP_002071972.1 | Bacterial Ig-like domain, group 2 | 2425 | 80 | 40 | 201.76 | 6.46 | UN/MS** |
| NP_714871.1 | Aconitatehydratase | 2358 | 97 | 44.5 | 82.3 | 6.38 | CY |
| WP_000565736.1 | Fructose-bisphosphatealdolase | 2348 | 66 | 67.8 | 38.18 | 6.46 | UN |
| NP_712306.1 | Periplasmic serine protease | 2180 | 49 | 36.3 | 36.68 | 8.85 | IN |
| NP_710494.1 | Elongation factor G | 2013 | 68 | 53 | 79.41 | 5.55 | CY |
| NP_711494.1 | Glutamine synthetase | 1952 | 74 | 48.2 | 53.42 | 5.9 | CY |
| YP_000947.1 | Outer membrane protein OmpL1 | 1878 | 68 | 66.6 | 33.55 | 8.69 | UN* |
| NP_710233.1 | NAD(P)(+) transhydrogenase subunit alpha | 1814 | 65 | 61.2 | 41.45 | 9.5 | IN |
| WP_000798840.1 | Serine protease | 1795 | 61 | 48.5 | 41.18 | 8.45 | PP* |
| NP_713600.1 | DNA-directed RNA polymerase subunit beta | 1775 | 76 | 42.5 | 138.01 | 5.6 | CY |
| WP_001062723.1 | Isocitrate dehydrogenase | 1742 | 72 | 46 | 44.9 | 6.53 | CY |
| NP_711909.1 | Hypothetical protein LA1728 | 1697 | 70 | 54.3 | 32.99 | 8.81 | CY* |
| NP_712247.1 | Hypothetical protein LA2066 | 1680 | 60 | 54.3 | 32.16 | 8.3 | UN/MS* |
Table 1: Top 30 proteins identified in pathogenic Leptospira OMVs by LC-MS/MS. A Subcellular location of each protein was predicted by PSORTb v3.0 http://www.psort.org/psortb/ CY: Cytoplasmic protein, IN: Inner/cytoplasmic protein, PP: Periplasmic protein, UN: Unknown location, UN/MS: Unknown location-This protein may have multiple localization sites). * Signal peptide type I is presented, predicted by LipoP 1.0 http://www.cbs.dtu.dk/services/LipoP/, ** Signal peptide type II is presented, predicted by LipoP 1.0 http://www.cbs.dtu.dk/services/LipoP/.
Humoral response induced by OMVs
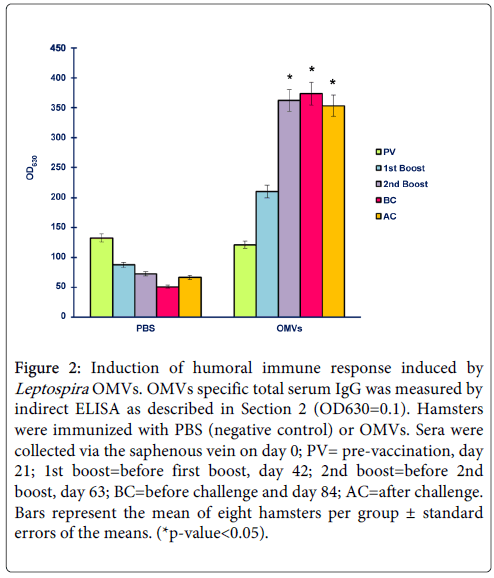
To assess whether OMVs were able to induce antibody responses specific to the antigens in vivo, two groups of hamsters were immunized 3 times at 3-week intervals with either PBS or 50 µg OMVs in the presence of adjuvants. Sera collected from hamsters, both control and vaccination group, at day 0 (pre-vaccination), day 21 (before 1st boost), day 42 (before 2nd boost), day 63 (before challenge) and day 84 (3 weeks after challenge) were measured for total IgG titers by indirect ELISA. As shown in Figure 2, L. interrogans OMVs induced high titers of OMVs-specific serum IgG after a single boost. It was clear that humoral responses necessary for battling the infection were raised significantly following immunization with OMVs, suggesting the antigenicity of OMVs. Increased antibody levels correlated with a high survival rate in vaccinated hamsters implying that antibody responses induced by OMVs contributed to protection.
Figure 2: Induction of humoral immune response induced by Leptospira OMVs. OMVs specific total serum IgG was measured by indirect ELISA as described in Section 2 (OD630=0.1). Hamsters were immunized with PBS (negative control) or OMVs. Sera were collected via the saphenous vein on day 0; PV= pre-vaccination, day 21; 1st boost=before first boost, day 42; 2nd boost=before 2nd boost, day 63; BC=before challenge and day 84; AC=after challenge. Bars represent the mean of eight hamsters per group ± standard errors of the means. (*p-value<0.05).
Protective efficacy of OMVs against challenge with virulent L. interrogans serovar Pomona in hamsters
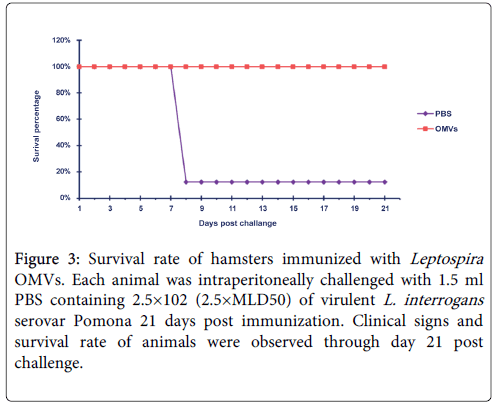
In addition to the ability of the vaccine candidate to stimulate antibody responses, protective efficacy is an important criterion for evaluation of vaccine potency; i.e. whether the antigen could protect animals against infection. To achieve this, all hamsters in the experiment were challenged with a single inoculum of the homologous Leptospira serovar. After challenge, all animals (8 of 8, 100%) in the vaccination group were protected, with 2 of them showing mild clinical signs of leptospirosis in the period of day 8 to 10 after challenge. Only one hamster (12.5%) in the control group survived until the end of experiment (3 weeks after challenge). This animal exhibited moderate signs of the disease, while the rest of them (7 of 8, 87.5%) suffered and died from acute leptospirosis on day 8 post-challenge (Figure 3). These results indicated that immunization of OMVs could provide protection upon lethal challenge.
Figure 3: Survival rate of hamsters immunized with Leptospira OMVs. Each animal was intraperitoneally challenged with 1.5 ml PBS containing 2.5×102 (2.5×MLD50) of virulent L. interrogans serovar Pomona 21 days post immunization. Clinical signs and survival rate of animals were observed through day 21 post challenge.
RT-PCR quantification of leptospire number
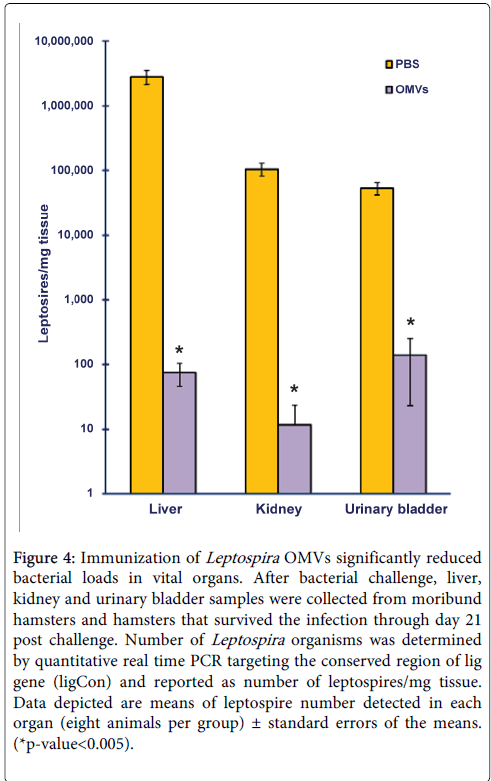
At the end of experiment, all surviving animals were euthanized and tissue samples were harvested from organs known to harbor leptospires--liver, kidney, and urinary bladder--for determination of bacterial load. In order to quantify more precise numbers of leptospires retained in these organs, real-time PCR using Taqman system, targeting the conserved region of lig gene, was used instead of the conventional culture method. The number of leptospires detected in liver, kidney and urinary bladder from hamsters immunized with OMVs were significantly fewer than those from the control group (p<0.005) (Figure 4). It is notable that for some hamsters immunized with OMVs, no leptospires weredetected in their kidney and urinary bladder samples, suggesting the ability of the immune system to recognize the spirochetes and clear the infection after immunization with OMVs (data not shown). To further confirm results from real time PCR, conventional culture in EMJH was done along with the assay. One month after media inoculation, none of the samples from OMV-immunized hamsters showed visible bacterial growth. On the other hand, all hamsters from the control group were Leptospira-positive except for one animal that survived the challenge (data not shown). In immunized hamsters, real time PCR detection of leptospires in tissues was positive, while it was negative when tissue samples were subjected to culture in EMJH. This could due to the fact that by the time tissues were harvested (21 days post infection), bacteria were inactivated by the immune system but not completely out of the body. Thus when total DNA was extracted from tissue samples, Leptospira gDNA was also isolated along with the animal’s DNA and was detected by real time PCR.
Figure 4: Immunization of Leptospira OMVs significantly reduced bacterial loads in vital organs. After bacterial challenge, liver, kidney and urinary bladder samples were collected from moribund hamsters and hamsters that survived the infection through day 21 post challenge. Number of Leptospira organisms was determined by quantitative real time PCR targeting the conserved region of lig gene (ligCon) and reported as number of leptospires/mg tissue. Data depicted are means of leptospire number detected in each organ (eight animals per group) ± standard errors of the means. (*p-value<0.005).
Histopathology examination of tissues
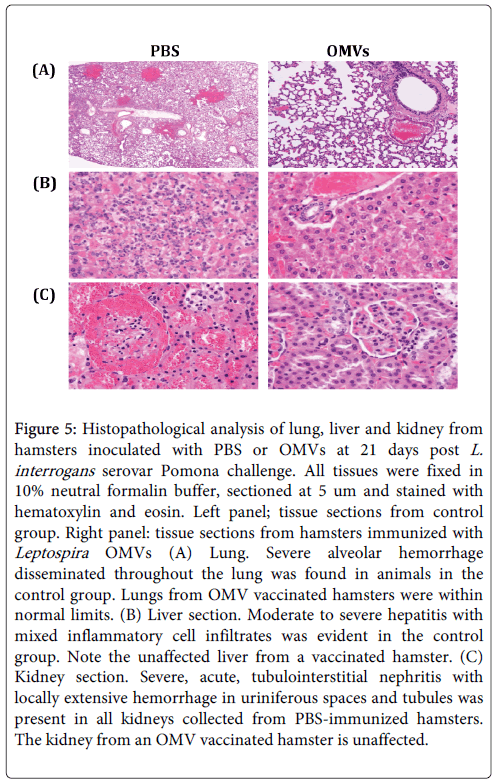
Histopathologic examination of various organs at 21 days after challenge demonstrated moderate to severe pulmonary lesions, hepatitis and hemorrhagic tubulointerstitial nephritis were predominant in PBS-immunized animals. Pulmonary lesions included thickening of alveolar septa by edema, interstitial leukocyte infiltration, endothelial cell swelling, and hemorrhage (Figure 5). In contrast, there were almost no inflammatory lesions present in the lung, liver or kidney from hamsters immunized with OMVs (70-80 % of animals had nolesions) (Table 2), consistent with the survival rate and leptospiral load, suggesting superior vaccine efficacy in induction of protective immunity.
Figure 5: Histopathological analysis of lung, liver and kidney from hamsters inoculated with PBS or OMVs at 21 days post L. interrogans serovar Pomona challenge. All tissues were fixed in 10% neutral formalin buffer, sectioned at 5 um and stained with hematoxylin and eosin. Left panel; tissue sections from control group. Right panel: tissue sections from hamsters immunized with Leptospira OMVs (A) Lung. Severe alveolar hemorrhage disseminated throughout the lung was found in animals in the control group. Lungs from OMV vaccinated hamsters were within normal limits. (B) Liver section. Moderate to severe hepatitis with mixed inflammatory cell infiltrates was evident in the control group. Note the unaffected liver from a vaccinated hamster. (C) Kidney section. Severe, acute, tubulointerstitial nephritis with locally extensive hemorrhage in uriniferous spaces and tubules was present in all kidneys collected from PBS-immunized hamsters. The kidney from an OMV vaccinated hamster is unaffected.
| Animal ID | Lung | Liver | Kidney | |||
|---|---|---|---|---|---|---|
| PBS | OMVs | PBS | OMVs | PBS | OMVs | |
| 1 | 3 | 0 | 3 | 1 | 3 | 0 |
| 2 | 1 | 0 | 2 | 0 | 3 | 1 |
| 3 | 2 | 0 | 2 | 1 | 3 | 0 |
| 4 | 2 | 0 | 3 | 0 | 3 | 0 |
| 5 | 2 | 0 | 3 | 0 | 3 | 1 |
| 6 | 2 | 0 | 3 | 0 | 3 | 0 |
| 7 | 3 | 0 | 2 | 0 | 3 | 0 |
| 8 | 0 | 0 | 0 | 0 | 0 | 0 |
| Average score | 1.875 | 0 | 2.25 | 0.25 | 2.625 | 0.25 |
Table 2: Prophylactic efficacy of Leptospira OMVs evaluated on the basis of histopathological lesions score in various organs. The pulmonary, hepatic and renal inflammatory lesions in hamsters were graded on a scale of severity; 0=within normal limit, 1=mild, 2=moderate, and 3=severe.
Discussion
Attempts to develop a safe and effective vaccine against leptospirosis have been made for years. Killed whole cell leptospiral vaccines produce protective responses that are serovar-specific, but outbreaks in different geographic areas usually involve different serovars [4]. Furthermore, side effects, including local and systemic reactions, have been reported [38]. For these reasons, a subunit vaccine based on protein antigens, especially outer membrane proteins, has been studied [19,32,39]. Considerable numbers of OMPs produced in the form of recombinant proteins have been tested for their immunogenicity and effectiveness [7,33,34]. However, the use of a single antigen from this group of proteins in vaccine trials could not provide complete protection when animals were challenged with either homo- or heterologous serovars [19,32]. Different vaccine strategies or a combination of multiple antigenic proteins in a single vaccine preparation may be required to yield maximum protection. Multicomponent vaccines based on several letpospiral OMPs [17] and heterologous prime-boost immunization strategy [18] have also been tried and are able to induce significant immunological responses.
The question is therefore raised whether OMVs produced from pathogenic L. interrogans are immunogenic and can induce protective immunity in animals against the challenge. OMVs are involved in multiple biological and physiological functions important for bacterial survival, biofilm production, quorum sensing, virulence and pathogenesis [25]. Electron microscopic examination of infected tissues and body fluids has demonstrated the presence of OMVs during infection. These vesicles contribute not only to bacterial colonization and host cell invasion, they also contain bacterial toxins and virulence factors [27]. Characterization of OMVs derived from other bacterial species and used in vaccine studies, showed that multiple well-established antigenic proteins, along with some virulence factors, were included in the vesicles [28-30,40]. Thus, OMVs contain surface antigens similar to those of killed whole cell or live-attenuated bacteria but are non-viable structures with fewer safety concerns.
Leptospira OMVs produced in the current study are considered non-native OMVs because they were produced by chemical treatment of the bacterial culture with low pH citrate buffer to detach the outer membrane from the protoplasm and then the cells were lysed. Ruptured outer membranes are able to close and become vesicles. It is interesting to note that, compared to natural OMVs harvested from cell culture supernatant, our vesicles contained a higher ratio of cytoplasmic protein. This may be because when the cells were lysed, non-secreted components, i.e. cytoplasmic or inner membrane proteins, were released into the mixture and encapsulated during vesicle formation. Even though OMVs extracted by this method are not in native form, nanoLC-MS/MS analysis of the vesicles identified multiple known leptospiral antigens (LipL32, LipL41 and OmpL1), some with high redundancy, together with other immunoreactive proteins (chaperonin [41,42], DnaK [41], elongation factor Tu, acetyl CoA acetyltransferase; Table 1). A similar result has been reported [43]. This data suggests Leptospira OMVs are a promising vaccine candidate. Moreover, production of OMVs could overcome the difficulties of expressing and purifying fused whole genes of various OMPs as multi-component vaccine. These features make the OMVs based vaccine more advantageous compared to conventional vaccine preparation, not only because multiple antigens are presented in the vesicles, but also due to the nature of their structures that helps maintain the native composition and conformation of the proteins which may contribute to a robust protective immune response, a response that may not be achieved when each antigen is administered alone.
Immunogenicity and protective efficacy of Leptospira OMVs were evaluated using a homologous challenge model in hamsters. We found that significantly increased levels of antibody generated by Leptospira OMVs confer protection, with a high survival rate, against homologous challenge with a highly virulent strain. It is well known that humoral immunity is the major immune effector responsible in combating the invasion of extracellular pathogens. Some studies [28,30] imply that an OMV vaccine may not need vaccine adjuvants because, besides the antigenic proteins, OMVs also contain pathogen-associated molecular patterns-PAMPs (lipoprotein, LPS, CpG DNA) in their natural contexts [27,44]. Binding of these innate immune-activating ligands to pattern recognition receptors (PRRs) would mimic those of an intact organism, which triggers proinflammatory cytokines and consequently enhances protective immune responses specific to the antigens. But since Leptospira is extracellular bacterium, bactericidal antibodies are required in order to clear the pathogen. Thus, the addition of vaccine adjuvant, e.g. Alum in order to drive the immune response towards humoral and Th2 [45], is suggested. Tissue samples from vaccinated animals were all culture negative suggesting that the induced immune response was able to either prevent colonization or aid in clearance of organisms.
In sum, immunization with OMVs not only protected the animal from lethal infection, but also helped decrease shedding of viable bacteria from infected animals into environment, which can be an important source of infection for humans and animals. Moreover, the presence of multiple antigenic OMPs that are highly conserved among pathogenic serovars and the capability to stimulate a strong immune response make OMV- based vaccines a very promising way to confer protection against challenge. In conclusion, this is the first study that provides a novel vaccine approach using Leptospira OMVs for the prevention of leptospirosis. Whether the OMVs can induce a cross protection against different serovars and/or induce a better protection than that of bacterin, further studies are warrants.
Acknowledgement
This work was supported in part by the Biotechnology Research and Development Corporation (BRDC). Financial supports for Anthicha Kunjantarachot were provided by Strategic Consortia for Capacity Building of University Faculties and Staff, Office of the Higher Education Commission, Bangkok, Thailand, 2012 Fulbright-Thailand Junior Research Scholarship Program and Interdisciplinary Graduate Program in Genetic Engineering, the Graduate School, Kasetsart University, Bangkok, Thailand. TEM image of Leptospira OMVs is courtesy of John L. Grazul at Cornell Center for Materials Research (CCMR) supported by NSF 1120296.
References
- Faisal SM, McDonough SP, Chang YF, editors. Chapter 8. Leptospira: Invasion, pathogenesis and persistence. M. E. Embers (Ed). The pathogenic spirochete: strategies for evasion of host immunity and persistence. New Yrok: Springer Science, 2012.
- Palaniappan RU1, Ramanujam S, Chang YF (2007) Leptospirosis: pathogenesis, immunity, and diagnosis. CurrOpin Infect Dis 20: 284-292.
- Katz AR1, Buchholz AE, Hinson K, Park SY, Effler PV (2011) Leptospirosis in Hawaii, USA, 1999-2008. Emerg Infect Dis 17: 221-226.
- Arent ZJ1, Andrews S, Adamama-Moraitou K, Gilmore C, Pardali D, et al. (2013) Emergence of novel Leptospiraserovars: a need for adjusting vaccination policies for dogs? Epidemiol Infect 141: 1148-1153.
- Silva EF, Cerqueira GM, Seyffert N, Seixas FK, Hartwig DD, et al. (2009) Leptospiranoguchii and human and animal leptospirosis, Southern Brazil. Emerg Infect Dis 15: 621-623.
- Vijayachari P1, Sugunan AP, Sharma S, Roy S, Natarajaseenivasan K, et al. (2008) Leptospirosis in the Andaman Islands, India. Trans R Soc Trop Med Hyg 102: 117-122.
- Thaipadungpanit J1, Wuthiekanun V, Chantratita N, Yimsamran S, Amornchai P, et al. (2013) Leptospira species in floodwater during the 2011 floods in the Bangkok Metropolitan Region, Thailand. Am J Trop Med Hyg 89: 794-796.
- Verma A1, Stevenson B (2012) Leptospiral uveitis - there is more to it than meets the eye! Zoonoses Public Health 59 Suppl 2: 132-141.
- Klopfleisch R1, Kohn B, Plog S, Weingart C, Nöckler K, et al. (2010) An emerging pulmonary haemorrhagic syndrome in dogs: similar to the human leptospiral pulmonary haemorrhagic syndrome? Vet Med Int 2010: 928541.
- Dwyer AE1, Crockett RS, Kalsow CM (1995) Association of leptospiralseroreactivity and breed with uveitis and blindness in horses: 372 cases (1986-1993). J Am Vet Med Assoc 207: 1327-1331.
- Brem S, Gerhards H, Wollanke B, Meyer P, Kopp H (1999) [35 leptospira isolated from the vitreous body of 32 horses with recurrent uveitis (ERU)]. Berl Munch TierarztlWochenschr 112: 390-393.
- Guerra MA (2013) Leptospirosis: public health perspectives. Biologicals 41: 295-297.
- Klaasen HL1, van der Veen M, Molkenboer MJ, Sutton D (2013) A novel tetravalent Leptospirabacterin protects against infection and shedding following challenge in dogs. Vet Rec 172: 181.
- Wilson S1, Stirling C, Thomas A, King V, Plevová E, et al. (2013) Duration of immunity of a multivalent (DHPPi/L4R) canine vaccine against four Leptospiraserovars. Vaccine 31: 3126-3130.
- Bharti AR1, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, et al. (2003) Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis 3: 757-771.
- Subharat S1, Wilson PR, Heuer C, Collins-Emerson JM (2012) Growth response and shedding of Leptospira spp. in urine following vaccination for leptospirosis in young farmed deer. N Z Vet J 60: 14-20.
- Feng CY, Li QT, Zhang XY, Dong K, Hu BY, (2009) Immune strategies using single-component LipL32 and multi-component recombinant LipL32-41-OmpL1 vaccines against leptospira. Braz J Med Biol Res 42: 796-803.
- Hartwig DD, Forster KM, Oliveira TL, Amaral M, McBride AJA, et al. (2013) A Prime-Boost Strategy Using the Novel Vaccine Candidate, LemA, Protects Hamsters against Leptospirosis. Clinical and Vaccine Immunology 20: 747-752.
- Chang YF1, Chen CS, Palaniappan RU, He H, McDonough SP, et al. (2007) Immunogenicity of the recombinant leptospiral putative outer membrane proteins as vaccine candidates. Vaccine 25: 8190-8197.
- Hartwig DD1, Seixas FK, Cerqueira GM, McBride AJ, Dellagostin OA (2011) Characterization of the immunogenic and antigenic potential of putative lipoproteins from Leptospirainterrogans. CurrMicrobiol 62: 1337-1341.
- Hauk P, Macedo F, Romero EC, Vasconcellos SA, de Morais ZM, Barbosa AS, et al. In LipL32, the major leptospiral lipoprotein, the C terminus is the primary immunogenic domain and mediates interaction with collagen IV and plasma fibronectin. Infect Immun 76: 2642-2650.
- Haake DA, Chao G, Zuerner RL, Barnett JK, Barnett D, et al. (2000) The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect Immun 68: 2276-2285.
- Holst J, Martin D, Arnold R, Huergo CC, Oster P, et al. (2009) Properties and clinical performance of vaccines containing outer membrane vesicles from Neisseria meningitidis. Vaccine 27 Suppl 2: B3-12.
- Nieves W, Asakrah S, Qazi O, Brown KA, Kurtz J, et al. (2011) A naturally derived outer-membrane vesicle vaccine protects against lethal pulmonary Burkholderiapseudomallei infection. Vaccine 29: 8381-8389.
- Kulp A, Kuehn MJ (2010) Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol 64: 163-184.
- Lee EY, Choi DS, Kim KP, Gho YS (2008) Proteomics in gram-negative bacterial outer membrane vesicles. Mass Spectrom Rev 27: 535-555.
- Ellis TN, Kuehn MJ (2010) Virulence and immunomodulatory roles of bacterial outer membrane vesicles. MicrobiolMolBiol Rev 74: 81-94.
- Roberts R, Moreno G, Bottero D, Gaillard ME, Fingermann M, et al. (2008) Outer membrane vesicles as acellular vaccine against pertussis. Vaccine 26: 4639-4646.
- Shang ES, Champion CI, Wu XY, Skare JT, Blanco DR, et al. (2000) Comparison of protection in rabbits against host-adapted and cultivated Borreliaburgdorferi following infection-derived immunity or immunization with outer membrane vesicles or outer surface protein A. Infect Immun 68: 4189-4199.
- Pierson T, Matrakas D, Taylor YU, Manyam G, Morozov VN, et al. (2010) Proteomic Characterization and Functional Analysis of Outer Membrane Vesicles of Francisellanovicida Suggests Possible Role in Virulence and Use as a Vaccine. Journal of Proteome Research 10: 954-967.
- Schild S, Nelson EJ, Bishop AL, Camilli A (2009) Characterization of Vibrio cholerae outer membrane vesicles as a candidate vaccine for cholera. Infect Immun 77: 472-484.
- Cao Y, Faisal SM, Yan W, Chang YC, McDonough SP, et al. (2011) Evaluation of novel fusion proteins derived from extracellular matrix binding domains of LigB as vaccine candidates against leptospirosis in a hamster model. 29:7379-7386.
- Nally JE, Whitelegge JP, Aguilera R, Pereira MM, Blanco DR, et al. (2005) Purification and proteomic analysis of outer membrane vesicles from a clinical isolate of LeptospirainterrogansserovarCopenhageni. Proteomics 5: 144-152.
- Yu NY, Wagner JR, Laird MR, Melli G, Rey S, et al. (2010) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26: 1608-1615.
- Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, et al. (2003) Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci 12: 1652-1662.
- Palaniappan RU, Chang YF, Jusuf SS, Artiushin S, Timoney JF, et al. (2002) Cloning and molecular characterization of an immunogenic LigA protein of Leptospirainterrogans. Infect Immun 70: 5924-5930.
- Palaniappan RU, Chang YF, Chang CF, Pan MJ, Yang CW, et al. (2005) Evaluation of lig-based conventional and real time PCR for the detection of pathogenic leptospires. Mol Cell Probes 19: 111-117.
- Koizumi N, Watanabe H (2005) Leptospirosis vaccines: past, present, and future. J Postgrad Med 51: 210-214.
- Palaniappan RU, McDonough SP, Divers TJ, Chen CS, Pan MJ, et al. (2006) Immunoprotection of recombinant leptospiral immunoglobulin-like protein A against Leptospirainterrogansserovar Pomona infection. Infection and immunity 2006 74: 1745-1750.
- Ayalew S, Confer AW, Shrestha B, Wilson AE, Montelongo M (2013) Proteomic analysis and immunogenicity of Mannheimiahaemolytica vesicles. Clin Vaccine Immunol 20: 191-196.
- Guerreiro H, Croda J, Flannery B, Mazel M, Matsunaga J, et al. (2001) Leptospiral proteins recognized during the humoral immune response to leptospirosis in humans. Infect Immun 69: 4958-4968.
- Natarajaseenivasan K, Artiushin SC, Velineni S, Vedhagiri K, Vijayachari P, et al. (2011) Surface-associated Hsp60 chaperonin of LeptospirainterrogansserovarAutumnalis N2 strain as an immunoreactive protein. Eur J ClinMicrobiol Infect Dis 30: 1383-1389.
- Sakolvaree Y, Maneewatch S, Jiemsup S, Klaysing B, Tongtawe P, et al. (2007) Proteome and immunome of pathogenic Leptospira spp. revealed by 2DE and 2DE-immunoblotting with immune serum. Asian Pac J Allergy Immunol 25: 53-73.
- Ellis TN, Leiman SA, Kuehn MJ (2001) Naturally Produced Outer Membrane Vesicles from Pseudomonas aeruginosa Elicit a Potent Innate Immune Response via Combined Sensing of Both Lipopolysaccharide and Protein Components. Infection and Immunity 78: 3822-3831.
- Lindblad EB (2004) Aluminium compounds for use in vaccines. Immunol Cell Biol 82: 497-505.
Copyright: © 2014 Kunjantarachot A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.