Indexed In
- RefSeek
- Hamdard University
- EBSCO A-Z
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Case Report - (2023) Volume 8, Issue 3
HNF1A Gene Mutation (c.811del, p.Arg271Glyfs) Causing Maturity Onset of Diabetes of the Young 3: A Case Study of an Indian Patient
Neelima Chitturi*, Satish Sunkara, Sandhya Kiran Pemmasani and Anuradha AcharyaReceived: 08-May-2023, Manuscript No. DCRS-23-21256; Editor assigned: 10-May-2023, Pre QC No. DCRS-23-21256(PQ); Reviewed: 30-May-2023, QC No. DCRS-23-21256; Revised: 06-Jun-2023, Manuscript No. DCRS-23-21256(R); Published: 13-Jun-2023, DOI: 10.35841/2572-5629.23.8.158
Abstract
Background: Maturity-Onset Diabetes of the Young (MODY), a rare class of diabetes that develops in early adulthood, is thought to have a prevalence of less than 5%. The best practice for the management of the MODY is to identify its type, through genetic testing. This paper presents a case study to identify the genetic etiology of diabetes.
Case history: The presented case was diagnosed with diabetes at the age of 17 with a strong family history of diabetes. The individual was on insulin (12 units) with high random blood sugar levels (200 mg/dL to 300 mg/ dL) and HbA1c (9.2%). Whole exome sequencing has shown the presence of a known likely pathogenic variant in the HNF1A gene (chr12: 121432062, c.811del, p.Arg271Glyfs), known to cause MODY3. Other unaffected family members were screened for the variant using Sanger sequencing, but the results were negative. The association of this variant with the disease is reported for the first time in the Indian population.
Conclusion: Blood sugar levels can be brought under control with the right medical care, such as sulphonylurea therapy, and eventually, painful insulin medication can be stopped. For a precise diagnosis and prognosis of diabetes, it's crucial to identify the genetic cause of the disease. This case demonstrates the critical role of genetic testing in proper treatment and management of diabetes.
Keywords
Case report; Diabetes; Genetics; HNF1A; MODY3; Sequencing
Introduction
Diabetes is of several types and Maturity-Onset Diabetes of the Young (MODY) is one among them. Its characteristics include hyperglycemia and persistent microvascular long-term consequences such as retinopathy, nephropathy, and neuropathy [1]. It is a relatively rare type of diabetes with an autosomal dominant mode of inheritance that typically manifests in adolescence or early adulthood, before the age of 25. It is estimated to have a prevalence of about 2% globally and 7.7% in India, which ranks second with highest number of diabetic patients after China [2-4]. There are several types of MODY, each caused due to mutations in different genes: MODY1 (HNF4A), MODY2 (GCK), MODY3 (HNF1A), MODY4 (PDX1), MODY5 (HNF1B), MODY6 (NEUROD1), MODY7 (KLF11), MODY8 (CEL), MODY9 (PAX4), MODY10 (INS), MODY11 (BLK), MODY13 (KCNJ11) and MODY14 (APPL1) [5, 6]. Thus, it is referred as a monogenic disorder. While many biochemical tests are used to determine the type of diabetes, whole exome sequencing helps to determine the type of diabetes, and to further plan the treatment strategy.
This study goes into great detail about the MODY3-causing mutation in the HNF1A gene. The HNF1A gene, which is located on chromosome 12, encodes a homodimeric protein that functions as a transcription factor for a number of genes unique to the pancreas and liver. It has a major role in the development of the liver and pancreas, glucose import and homeostasis, insulin production, and renal glucose absorption [6, 7]. Researchers found up to 350 distinct mutations in the gene's promoter region and exons in diabetic patients, and a few have also been found in Indians [1, 8 - 11]. The objective of this study is to demonstrate the importance of identifying underlying genetics of diabetes through whole exome sequencing.
Case Presentation
Clinical History
The patient was diagnosed with diabetes at the age of 17 years and was on insulin (12 units). The random blood sugar levels were 200 mg/dL to 300 mg/dL and HbA1c was 9.2%. The patient was recommended for MODY testing after ruling out the possibilities of type 1 and type 2 diabetes. The parents of the proband are healthy and non-diabetic. Their random blood sugar levels and HbA1c were within normal limits. The pedigree is given in Figure 1.
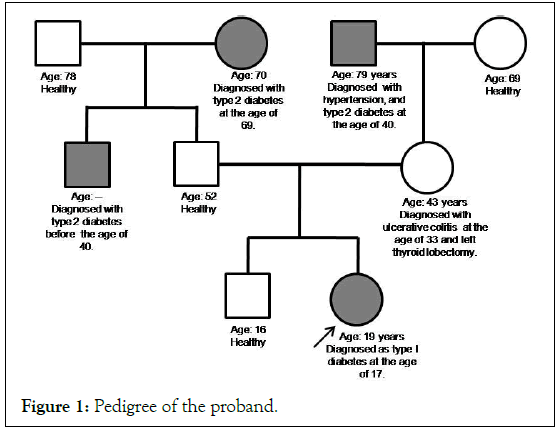
Figure 1: Pedigree of the proband.
All experimental and analytical procedures were performed as per standard manufacturer’s protocol. An informed written consent mentioning the use of the sample for research was taken from the proband and her family members. Research was carried out in compliance with the Helsinki declaration and the procedures had been approved by internal bio-safety committee at Mapmygenome.
Extraction of DNA from blood
Genomic DNA was extracted from the blood samples using Nucleospin DNA isolation kit from Takara Bio and quantified spectrophotometrically using QubitTM from Thermo Fisher Scientific.
Whole exome sequencing and analysis
Exome libraries were prepared using Twist Comprehensive Exome Panel, from Twist Bioscience. Sequencing was performed on a NovaSeq 6000 Illumina sequencer and paired-end reads of length 150 bases were generated. Quality of raw data was verified using fastqc tool version V0.1.1 and adapters were removed using Trimommatic software version 0.39. The trimmed paired end reads were aligned to reference human genome (hg19 version) using BWA version 0.7.17. Haplotype Caller package, of GATK version 4.2.0.0, was used to call variants as per Broad Institute’s Best Practices protocol. Variants were annotated using VEP release 104 (against the Ensembl release 75 human gene model) and data from other resources such as dbsnp, dbnsfp, snpeff, ClinVar, HGMD, OMIM and HPO. The variants were filtered using criteria: a) variants of genes associated with diabetes in general and MODY in particular b) exclude synonymous, 5’ and 3’variants c) variants with gnomAD & 1000Genomes frequency less than 5%. Variant classification was done as per American College of Medical Genetics and Genomics (ACMG) guidelines [12].
Targeted validation by Sanger sequencing
The presence or absence of variant was confirmed in samples of mother and brother using bidirectional Sanger sequencing. Father was not available and so his sample could not be tested for the identified variant.
Results and Discussion
Whole exome sequencing of the proband revealed a heterozygous frame- shift deletion in exon 4 of HNF1A gene (NM_001306179.2:c.811del, non-functional gene product and leads to nonsense mediated decay, which is an established disease mechanism (ACMG: PVS1).
This particular variation is not seen in her mother and her sibling (Figure 2) and is likely a spontaneous mutation in the proband (ACMG: PM6). The variant is not found in gnomAD population database (ACMG: PM2). Variants in other genes that are known to cause diabetes were also considered for analysis, and no other significant variants were found (ACMG: PP4). The variant was classified by ClinVar as Likely Pathogenic (ACMG: PP5) based on unpublished results [13]. Based on above ACMG criteria, we recommend this particular variant to be re-classified as ‘Pathogenic’ for MODY3.
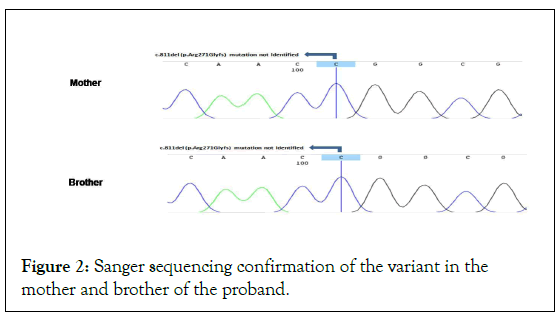
Figure 2: Sanger sequencing confirmation of the variant in the mother and brother of the proband.
Various types of mutations in HNF1A gene have been identified in diabetic patients [8 -11]. The most common mutations are c.872dupC in exon 4 (present in 234 affected families) and c.391C>T in exon 2 (present in 29 affected families) [14]. But the variant reported in this case study has not been reported in the Indian population to the best of our knowledge. Mutations in the HNF1A gene can alter the normal functioning of the pancreas, impairing insulin synthesis and leading to high blood sugar levels. Affected people have hyperglycemia and a higher chance of developing other issues, such as diabetic nephropathy and cardiovascular problems. Patients with HNF1A mutations have been observed to respond particularly well to sulfonylurea, according to research. Its use can postpone the requirement for insulin therapy and, over time, lessen microvascular problems [15]. Though the current study could identify the genetic cause of diabetes it is limited in terms of lack of follow-up on the patient's response to sulfonylurea therapy.
Conclusion
According to 2019 predictions, 77 million Indians have diabetes, and by 2045, that number is projected to rise to 134 million. Finding the genetic basis of diabetes is crucial for effective treatment and a better prognosis. Correct therapy at the earliest phases of the illness will result in fewer health issues and better management. The case study presented here strongly supports the need for genetic testing for diabetic therapy.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Conflict of Interest
The authors declare no potential conflicts of interest.
Author Contributions
Material preparation and wet lab analysis was performed by (Satish Sunkara), pipeline development by (Sandhyakiran Pemmasani), in silico analysis and variant identification by (Neelima Chitturi), the first draft of the manuscript was written by (Neelima Chitturi) and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Compliance with Ethical Standards and Ethical Approval
An informed written consent mentioning the use of the sample for research was taken from the participants. Research was carried out in compliance with the Helsinki declaration and the procedures have been approved by internal bio-safety committee at Mapmygenome.
Funding
This study was not funded by any agency.
Consent to Participate and/or Consent to Publish
Informed written consent was obtained from all individual participants included in the study. The authors affirm that human research participants provided informed consent for publication of the image in Figure 2.
References
- Valkovicova T, Skopkova M, Stanik J, Gasperikova D. Novel insights into genetics and clinics of the HNF1A-MODY. Endocr Regul. 2019;53(2):110-134.
[Cross Ref] [Google Scholar] [PubMed]
- Kim SH. Maturity-onset diabetes of the young: what do clinicians need to know?. Diabetes & metabolism journal. 2015;39(6):468-477.
[Cross Ref] [Google Scholar] [PubMed]
- Bhat JA, Masoodi SR, Bhat MH. Prevalence and clinical profile of maturity onset diabetes of the young among people with diabetes attending a tertiary care institute in North India. Indian Journal of Endocrinology and Metabolism. 2022;26(Suppl 1):S25.
- Saeedi P, Petersohn I, Salpea P, Malanda B, Karuranga S, Unwin N, et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas. Diabetes research and clinical practice. 2019;157:107843.
[Cross Ref] [Google Scholar] [PubMed]
- Oliveira SC, Neves JS, Pérez A, Carvalho D. Maturity-onset diabetes of the young: from a molecular basis perspective toward the clinical phenotype and proper management. Endocrinologia, diabetes y nutricion. 2020;67(2):137-147.
[Cross Ref] [Google Scholar] [PubMed]
- Fajans SS, Bell GI, Polonsky KS. Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. N Engl J Med. 2001;345(13):971-980.
[Cross Ref] [Google Scholar] [PubMed]
- Hoffman LS, Fox TJ, Anastasopoulou C, Jialal I. Maturity Onset Diabetes in the Young. StatPearls Publishing. 2022.
[Cross Ref] [Google Scholar] [PubMed]
- Radha V, Ek J, Anuradha S, Hansen T, Pedersen O, Mohan V. Identification of novel variants in the hepatocyte nuclear factor-1α gene in South Indian patients with maturity onset diabetes of young. J Clin Endocrinol Metab. 2009;94(6):1959-1965.
[Cross Ref] [Google Scholar] [PubMed]
- Plengvidhya N, Tangjittipokin W, Teerawattanapong N, Narkdontri T, Yenchitsomanus PT. HNF1A mutation in a Thai patient with maturity-onset diabetes of the young: A case report. World J Diabetes. 2019;10(7):414.
[Cross Ref] [Google Scholar] [PubMed]
- Balamurugan K, Bjorkhaug L, Mahajan S, Kanthimathi S, Njolstad PR, Srinivasan N, et al. Structure–function studies of HNF1A (MODY3) gene mutations in South Indian patients with monogenic diabetes. Clin Genet. 2016;90(6):486-495.
[Cross Ref] [Google Scholar] [PubMed]
- Miyachi Y, Miyazawa T, Ogawa Y. HNF1A mutations and beta cell dysfunction in diabetes. Int J Mol Sci. 2022;23(6):3222.
[Cross Ref] [Google Scholar] [PubMed]
- Masson E, Zou WB, Genin E, Cooper DN, Le Gac G, Fichou Y, et al. Expanding ACMG variant classification guidelines into a general framework. Hum Genomics. 2022;16(1):1-5.
[Cross Ref] [Google Scholar] [PubMed]
- Ellard S, Colclough K. Mutations in the genes encoding the transcription factors hepatocyte nuclear factor 1 alpha (HNF1A) and 4 alpha (HNF4A) in maturity‐onset diabetes of the young. Hum Mutat. 2006;27(9):854-869.
[Cross Ref] [Google Scholar] [PubMed]
- Colclough K, Bellanne‐Chantelot C, Saint‐Martin C, Flanagan SE, Ellard S. Mutations in the genes encoding the transcription factors hepatocyte nuclear factor 1 alpha and 4 alpha in maturity‐onset diabetes of the young and hyperinsulinemic hypoglycemia. Human mutation. 2013;34(5):669-685.
[Cross Ref] [Google Scholar] [PubMed]
- Bacon S, Kyithar MP, Rizvi SR, Donnelly E, McCarthy A, Burke M, et al. Successful maintenance on sulphonylurea therapy and low diabetes complication rates in a HNF1A–MODY cohort. Diabet Med. 2016;33(7):976-984.
[Cross Ref] [Google Scholar] [PubMed]
- Pradeepa R, Mohan V. Epidemiology of type 2 diabetes in India. Indian J Ophthalmol. 2021;69(11):2932.
[Cross Ref] [Google Scholar] [PubMed]
Citation: Chitturi N, Sunkara S, Pemmasani SK, Acharya A (2023) HNF1A Gene Mutation (c.811del, p.Arg271Glyfs) Causing Maturity Onset of Diabetes of the Young 3: A Case Study of an Indian Patient. Diabetes Case Rep. 8:158.
Copyright: © 2023 Chitturi N, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
