Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- CiteFactor
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2020) Volume 11, Issue 9
Identification and Characterization of Bacterial Agents Causing Moderate Damage and Souring of the Fig Fruits
Seyedeh Asiyeh Mousavi1*, Nader Hasanzadeh1 and Parisa Abdollahi22Department of Agricultural Biotechnology, Faculty of Agriculture and Food Industries,Islamic Azad University, Tehran, Iran
Received: 31-Jul-2020 Published: 16-Sep-2020, DOI: 10.35248/2157-7471.20.11.514
Abstract
In order to determine the factors of decay and sourness of Fig fruits, in the summer of 2016-17, 60 leaf, fruit and stem samples from different regions of Tehran, Varamin (Qal'e No), Mazandaran (Amol, Noor and Sari), Lorestan and a branch of Fig fruit sample from Italy were collected. The samples were surface disinfected with 70% alcohol, washed and macerated in DH2O and one loopful of each suspension was cultured on NA and KB media. The dominant colonies from each culture were further selected and purified. The pathogenicity tests were fulfilled by Hypersensitivity reaction on geranium leaves and artificially by Fig fruit inoculation. Thirty isolates were considered as pathogenic bacteria and key phenotypic traits were performed. This was confirmed by 16S rRNA gene PCR assay using a pair of universal P1/P6 primer. The amplified PCR products were sequenced by Macrogene Inc. and were Blast in NCBI database (www.ncbi.nlm.nih.gov). The bacteria, Stenotrophomonas maltophilia, Pseudomonas aeruginosa, Pseudomonas fulva, Brevibacterium linens, Pseudomonas fragi, Bacillus licheniformis, Bacillus paralicheniformis and Bacillus cereus were identified. None of the isolates caused fruit rot but typical disease symptoms were observed on Fig leaves and fruits. This is the first report of the presence of pathogenic bacteria on Fig trees in Iran.
Keywords
Fig trees; Bacteria; Iran; 16S rRNA gene; Sequencing
Introduction
Among the agricultural products, fruits are the most important horticultural products that play an important role in supplying human nutrition and health. Among these, the Fig (Ficus carica L.) is one of the earliest cultivated fruit trees in the world [1]. The tree is cultivated in warmest and temperate Mediterranean climates with fifty percent of the world’s fig production concentrated in the Mediterranean area. Fresh and dried figs are consumed and appreciated world-wide not only for their unique taste and distinct flavor, but also for their health benefits. Figs have been popular not only because of their pleasant taste but possibly also because of their medicinal properties. The short postharvest life of fresh fruits is a critical point for the marketability in respect to dried fruits. The fresh figs are highly perishable fruits being susceptible to postharvest deterioration, such as softening, bruising, splitting, and pathogens growth. In Iran, Fars province and Estahbanat city are considered as the poles of Fig production [2]. According to the Agricultural Jihad statistics, 86685.5 hectares of dry and rainfed agricultural lands of the country were allocated to the commercial Fig production [3]. FAO statistics also show that Iran's rank in terms of production and cultivation of Fig is fifth and third, respectively [4]. Many biotic factors have been forced to reduce the production of Figs in gardens and in warehouses. These include the fungi, Cercospora leaf spot (Cercospora fici), Fig souring (Hanseniospora osmophila), Fig brown rot (Fusarium moniliforme) and Fig mosaic virus (unknown virus) [2,5] Some other Fig fungal diseases include Botrytis blight (Botrytis cinerea), Thread blight (Pellicularia koleroga) results in necrosis of stems and matted foliage, Botryosphaeria dothidea which cause necrosis of leaves and stems and Rhizopus stolonifer (smut) as the causal of fruit drop of cultivars with an open eye. Fusarium spp. and Aspergillus niger are also attack ripe fruits [6].
Few bacterial diseases were also reported on Ficus spp. particularly on edible Figs (Ficus carica). These include crown gall (Agrobacterium tumefaciens), Pseudomonas leaf spot (Pseudomonas cichorii) and Xanthomonas leaf spot caused by Xanthomonas campestris pv. fici [7,8]. Rhizobium larrymoorei (=A. larrymoorei) was isolated from gall of weeping Fig (Ficus benjamina) [9].
Apart these, various yeasts, fungi, and bacteria apparently responsible for the souring disease of the fruit of the Fig. Organisms are carried into the fruit by the dried fruit beetle, Carpophilus hemipterus. It is a preharvest problem resulting from yeasts and bacteria carried into the Figs by insects, especially vinegar flies, resulting in odors of alcohol or acetic acid [10-12]. Many types of Fig fruit rot were observed, from the soft, watery, fermented type to the typical dryrot type [13]. The aim of this study was to identify new bacterial pathogens and some bacteria affecting fruit souring of Fig trees.
Materials and Methods
Isolation and purification of bacteria
In the summer and late autumn of 2016, 60 plant samples including branch, leaf and fruit of infected Fig trees with brown spot symptoms on leaves and rot and deformity of fruits and necrotic spots on fruit and stems were collected from different regions of Tehran, Mazandaran and Varamin. A peculiar Fig samples were collected from a tree grown in Modarres hospital yard in Tehran. All samples were washed with distilled water and then sterilized with 70% alcohol for 1 minute. Small pieces from border of infected and healthy tissues were prepared and each was transferred to tubes containing 5 ml sterilized distilled water. A loopful of sample suspension was streaked on Nutrient agar (NA) and King's (KB) media. Petriplates were incubated at 25°C for 48 hours. The typical single colonies were transferred to new culture media and purified. Pathogenicity tests were fulfilled and all suspected pathogens were preserved in nutrient broth containing 15% glycerol at -20°C and in NA slants under liquid paraffin.
Pathogenicity tests
Hypersensitivity reaction (HR): The hypersensitivity reaction test was performed on geranium leaves. At first, a suspension of bacteria with an approximate concentration of 106 cfu/ml was prepared and then injected with a sterile syringe to underside of the young leaves. The reaction was evaluated for 24 to 48 hours [14].
Potato soft rot test: Healthy and fresh potato tubers were washed thoroughly with tap water, surface sterilized with 70% alcohol. The tuber skin was peeled and internal tissue was cut into small slices. Each slice was placed into sterile petri dish containing about 1/8 inch of sterile water and the surface was of the tissues were inoculated with a single colony of each isolate. Distilled water was used as negative controls. Petri dishes were kept at 25-30°C for 24- 24 hours. All rotten and decay slices were recorded [15].
Pathogenicity test on detached Fig fruits and leaves: The fruits and leaves of two Fig varieties (desert King Fig and Adriyatic Fig) were first disinfected with alcohol and then were put into sterilized petri dishes containing sterile distilled water. The leaves and Fig fruits were inoculated with individual bacterial suspension using straight wire [15]. Distilled water is used as a negative control. The negative treatments were checked daily for development of any disease symptoms.
Phenotypic characterization of the isolates
The isolates were characterized based on certain key tests. These include, gram test, aerobic/anaerobic growth, NaCl tolerance, spore formation, fluorescent pigmentation, levan production, oxidase, catalase, gelatinase, amylase, β-glucosidase, urease and nitrate reductase tests. Citrate utilization, casein and tween hydrolyses, MR-VP tests were also tested [15].
Identification based on PCR analysis
DNA extraction: Bacterial DNA was extracted by simple boiling
method modified after Sambrook et al. [16]. Bacteria were grown in agar media at 28°C for 2 days. A loopful of cells was suspended in 25 μl of sterile distillated water and followed an addition of 25 μl of freshly prepared lysis buffer containing 0.1 N NaOH and 0.5% SDS. The mixture was boiled in a water bath for 15 min. 200 μl of TE (10 mM Tris-HCl pH 8.0, 0.1 mM EDTA) was added to the mixture and centrifuged at 13,000 rpm for 5 minutes. Each supernatant was transferred to a clean tube and stored at -20 °C.
Preparation of mixture reaction for PCR test: The final volume of the reaction mixture was 25 μl for each tube, containing 23 μl of the reaction mixture and 1 μl of genomic DNA equivalent to 100 ng for each isolate. The primer pair P1 (5′-AGAGTTTGATCCTGGTCAGAACGCT-3′) and P6 (5′-TACGGCTACCTTGTTACGACTTCACCCC-3′) was used [17]. The reaction mix was placed on a thermocycler and subjected to PCR cycles: 94°C for 2 min, followed by 94°C for 45 seconds, annealing at 58°C for 45 seconds, primer extension at a temperature of 72°C for 1 minute and final extension by 35 cycles at a temperature of 72°C for 20 minutes. PCR amplified fragments were electrophoresed in an agarose gel (1%) and were visualized using loading buffer dye. PCR products of 10 strains were sent to Macrogen Inc. (Seoul, South Korea) for sequencing. The DNA sequences are compared with existing sequences in the NCBI GenBank. Phylogenetic tree from aligned sequences was constructed using Mega-6 software.
Results
Isolation and characterization of the isolates
Sixty Fig plant samples with different diseased and abnormal symptoms from 7 cities and one from Italy were collected (Figure 1). Thirty bacterial isolates were selected in the first stage that were capable of causing HR on tobacco leaves and to some extent disease symptoms of Fig leaf and fruit. Of these, 10 isolates with sever disease symptoms were selected for PCR analysis.
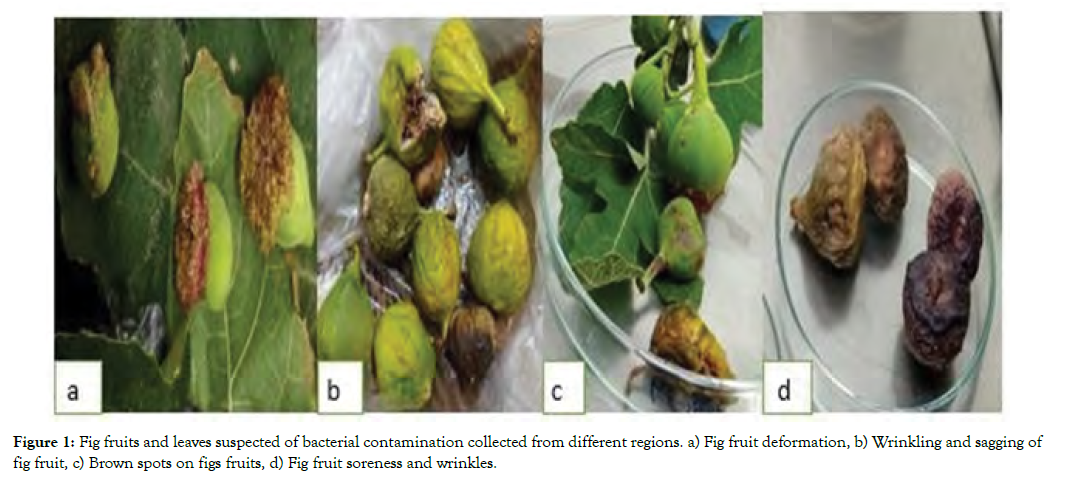
Figure 1: Fig fruits and leaves suspected of bacterial contamination collected from different regions. a) Fig fruit deformation, b) Wrinkling and sagging of fig fruit, c) Brown spots on figs fruits, d) Fig fruit soreness and wrinkles.
Pathogenicity tests
Hypersensitivity reaction: Thirty isolates were able to cause hypersensitivity and necrosis on pelargonium leaves (Figure 2).
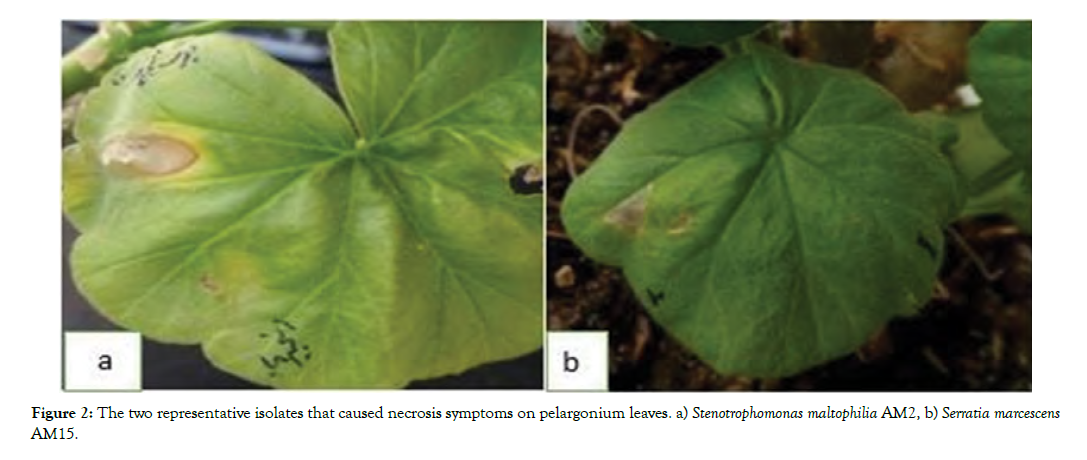
Figure 2: Incidence of necrosis and chlorosis symptoms on leaves and fruits by isolates a) Stenotrophomons maltophilia AM2, b) Pseudomonas aeruginosa AM22, c) Brevibacterium linens AM12.
Potato soft test: None of the 30 isolates that showed necrosis on the pelargonium leaves were able to soft rot on the potato slices.
Testing for pathogenicity on Fig fruits and leaf: All 10 selected isolates caused various disease symptoms including necrosis and chlorosis in leaves; rot and deformity of fruits and brown spots on fruit skin and stem. The four bacteria namely, Pseudomonas fragi, Brevibacterium linens, Serratia marcescens, Pseudomonas aeruginosa could have severe symptoms of the disease on Adriatic fruits rather than desert king Fig fruits (Figure 3).
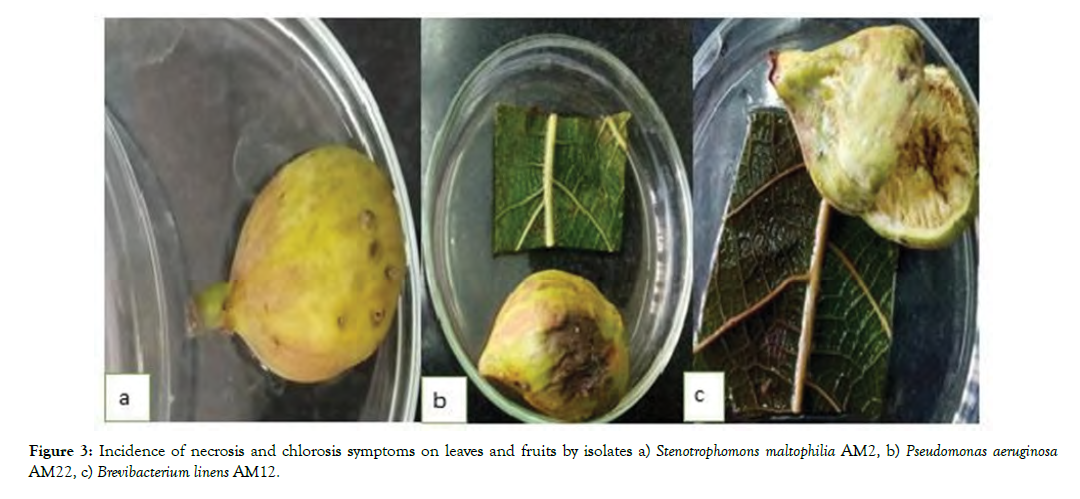
Figure 3: Incidence of necrosis and chlorosis symptoms on leaves and fruits by isolates a) Stenotrophomons maltophilia AM2, b) Pseudomonas aeruginosa AM22, c) Brevibacterium linens AM12.
Identification of bacterial isolates
The ten selected bacteria were identified based on certain morphological, physiological and biochemical tests (Table 1).
| Isolates/Characteristic | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Gram reaction | - | + | - | + | + | - | - | - | - | + |
| Colony color | Y | W | W | C | W | W | Y | M | W | W |
| Fluorescentpigment on KB | + | - | + | - | - | - | - | + | - | - |
| O/F test | O | F | O | O | F | F | O | O | F | F |
| Catalase | + | + | - | + | + | + | - | + | + | + |
| Nitratereduction | + | + | - | - | + | + | - | + | + | + |
| Oxidase | - | - | + | + | - | - | + | + | - | - |
| Argininedihydrolase | - | + | - | + | + | + | - | + | + | + |
| Hydrolysis: | ||||||||||
| Starch | - | + | - | - | + | - | - | + | - | + |
| Gelatin | + | - | - | + | - | + | - | + | + | - |
| Tween 80 | + | - | + | - | - | + | - | + | + | - |
| Levan production | - | - | - | - | - | - | - | - | - | - |
| Potato soft rot | - | - | - | - | - | - | - | - | - | - |
| HR on geranium | + | + | + | + | + | + | + | + | + | + |
Table 1: Physiological and biochemical tests for differentiating 10 selected fig fruits. 1) Stenotrophomons maltophilia AM2; 2) Bacillus licheniformis AM 5; 3) Pseudomonas fragi AM11; 4) Brevibacterium linens AM12; 5) Bacillus paralicheniformis AM13; 6) Serratia marcescens AM15; 7) Pseudomonas fulva AM18; 8) Pseudomonas aeruginosa AM22; 9) Serratia marcescens AM26; 10) Bacillus cereus AM27; M: Milky; Y: Yellow; C: Creamy; W: White; O/F; O: Aerobe; F: Facultative anaerobe.
Determination of the 16S rDNA gene Sequences
All 10 selected bacteria produced a single sharp 1500bp fragment (Figure 4). The PCR products were sent to the Macrogene Inc for sequencing. Sequencing was done in both directions of Sense and Antisense using P1/P6 primers. The sequence alignments were done using BLAST software in NCBI database (www.ncbi.nlm.nih. gov). The results were indicated that the strains AM2, AM5 and AM11 by 100% of homology were belonged to Stenotrophomonas maltophilia, Bacillus paralicheniformis and Pseudomonas fragi, respectively. Likewise, the strains AM13, AM15 and AM26, AM22 and AM27 by 100% of homology were assigned as Bacillus licheniformis, Serratia marcescens, Pseudomonas aeruginosa and Bacillus cereus, respectively. Percentage of homology 96% and 84% ensured that The 16S rRNA gene sequences of the strains AM12 and AM18 were also showing 96% and 84% identity to Brevibacterium linens and Pseudomonas fulva, respectively.
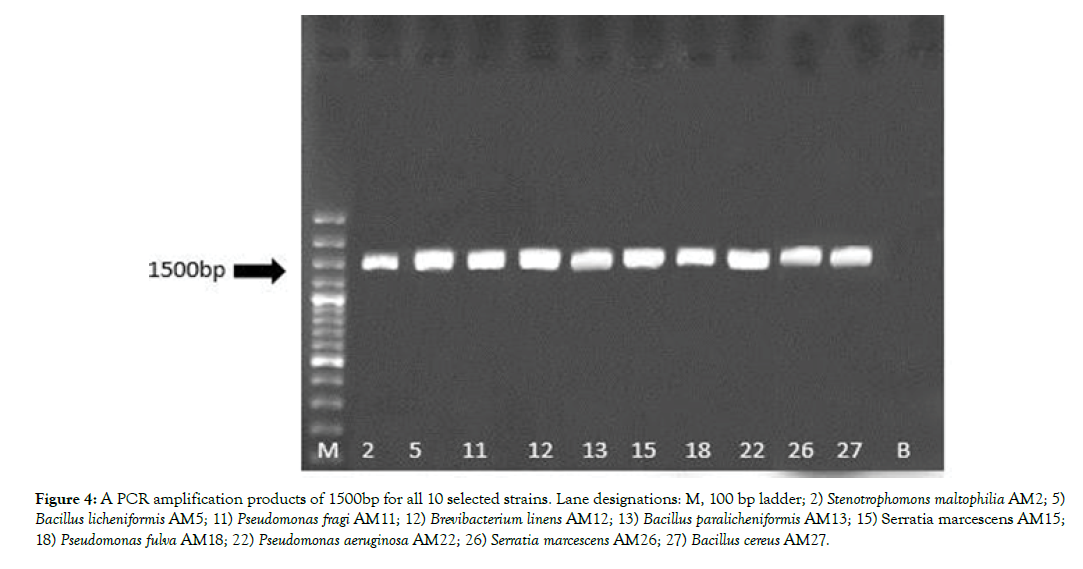
Figure 4: A PCR amplification products of 1500bp for all 10 selected strains. Lane designations: M, 100 bp ladder; 2) Stenotrophomons maltophilia AM2; 5) Bacillus licheniformis AM5; 11) Pseudomonas fragi AM11; 12) Brevibacterium linens AM12; 13) Bacillus paralicheniformis AM13; 15) Serratia marcescens AM15; 18) Pseudomonas fulva AM18; 22) Pseudomonas aeruginosa AM22; 26) Serratia marcescens AM26; 27) Bacillus cereus AM27.
Their GenBank accession numbers of the strains are as follows:
MH001477, MH012180, MH012187, MH012195, MH012196, MH012198, MH013188, MH014944, MN461457 and MN461460.
Phylogenetic analysis
Phylogenetic tree based on the 16S rRNA gene of 10 pathogenic bacteria of Fig plants (Figure 5). The evolutionary history was inferred by using the Maximum Likelihood method based on the Kimura 2-parameter model. The numbers at the nodes are bootstrap values based on 1000 replications. Evolutionary analyses were conducted in MEGA-6 [18].
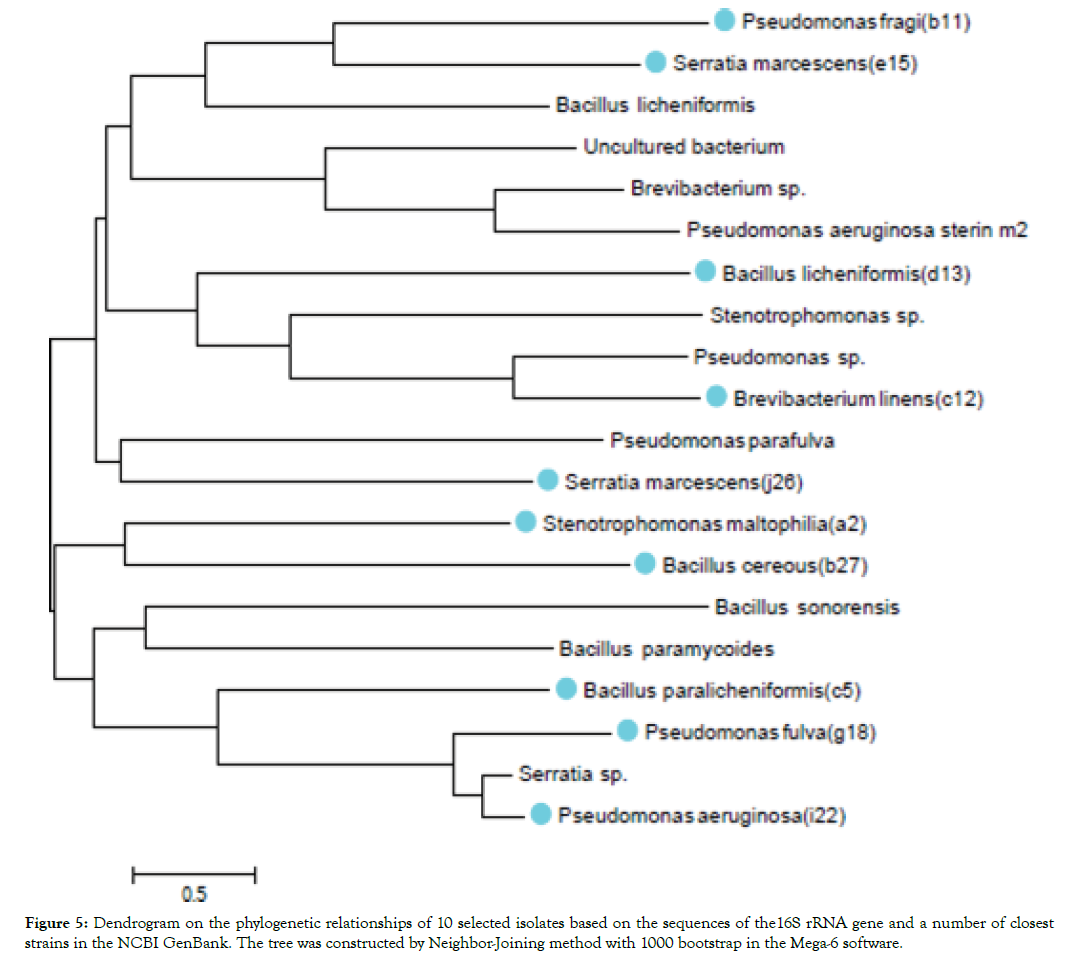
Figure 5: Dendrogram on the phylogenetic relationships of 10 selected isolates based on the sequences of the16S rRNA gene and a number of closest strains in the NCBI GenBank. The tree was constructed by Neighbor-Joining method with 1000 bootstrap in the Mega-6 software.
Discussion
Despite the importance of Fig for Iran, which is economically valuable, the bacterial diseases of the Fig trees, has remained silent until now. Perhaps one of the reasons is that there are a few reports of bacterial diseases on this host worldwide. Most diseases are caused by fungi and most prevalent fruit infection is souring which has been caused by various yeasts, fungi, and bacteria, carried into the fruit by the dried fruit beetle, Carpophilus hemipterus [8,13]. In this research, it was attempted to collect all contaminated samples from gardens of seven cities and one from the hospital yard and one from Italy in order to isolate some pathogenic bacteria. Overall of 60 samples were collected, of which 30 isolates were cultured and purified. 10 isolates with severe virulence were selected and further analyzed by PCR method. Pathogenicity tests were included HR on geranium leaves, potato soft rot and direct injection of bacterial suspensions into leaf midribs and immature Fig fruits. Fruit skins were pierced by needles infected with bacterial colonies. Necrosis and water-soaking were the two characteristic leaf symptoms were observed. Fruit symptoms varied from discoloration and decay in the internal tissue of Fig fruits to brown spots on the fruit skins.
Apart these, Pseudomonas aeurginosa AM22 caused the wrinkling of the fruits, as it had been seen in natural conditions. On the other hand, Serattia marcescens AM15 and AM26, Brevibacterium linens AM12, Stenotrophomonas maltophilia AM2 and Pseudomonas fragi AM11, caused brown spots, water-soaking lesions and fruit spoilage. The three more isolates namely Bacillus licheniformis AM13, Pseudomonas fulva AM18 and Bacillus paralicheniformis AM5 could cause brown spots and water-soaking lesions without fruit spoilage.
All bacteria were negative to potato soft rot test. This indicates that their pathogenic potency was not dependent on the activity of the cellulolytic and pectiolytic enzymes. All isolates were identified on the basis of biochemical traits and then confirmed by PCR analysis using P1/P6 as a universal primer pair for amplification of 16S rRNA gene.
The distribution of these bacteria and the areas where these isolates were isolated are as follows: P. aeruginosa and S. marcescens were isolated from leaf and fruits of the Fig trees located in Modarres hospital in Tehran. P. fragi and S. maltophilia were isolated from leaves and ripe fruits of Fig trees in green space of Varamin Research Institute, respectively.
S. maltophilia and B. paralicheniformis were isolated from ripe fruits and leaves of Figs trees in Varamin city, respectively. P. fulva from leaf and B. linens from fruit samples of Amol city. B. licheniformis was the only isolate which was collected from Eram Park-Tehran.
Among these, B. cereus was the only isolate obtained from a single Fig sample of the Bari-Italy. An important finding in this study was the isolation of Serattia marcescens, Stenotrophomonas maltophilia, Pseudomonas aeruginosa, Pseudomonas fulva, Bacillus licheniformis, Bacillus cereus as common bacteria in humans and plants from Fig trees in different regions.
According to numerous reports, these bacteria cause an infection in humans, especially in patients with immune deficiency [19,20]. It was reported by Kamar, 1989 that Bacillus cereus is the cause of two types of poisoning (diarrhea and vomiting). In the case of Brevibacterium linens and Pseudomonas fragi, these two species have been isolated from plant and dairy products and have been caused the consumer illness [21] whereas; Pseudomonas fragi was isolated from lettuce [22]. Pseudomonas aeruginosa, a common opportunistic bacterium between humans and plants was isolated from diferent sources such as rhizosphere of corn [23] and tobacco leaf in China [24]. Another important species was Pseudomonas fulva, a clinical isolate which has been reported mostly from aquatic environments including rice seed and fennel plants [22]. Serattia marcescens, a ubiquitous bacterium has been isolated from plant materials such as watermelon and nutty trees [25]. The latter, was first isolated from Figs in New York, USA [22].
Bacillus licheniformis, has a wide range of hosts and is isolated from soil containing vegetable waste [26], wheat fields of central provinces, Golestan and Mazandaran [27], cucumber farrowing in Jiroft [28], soil and water around the hot springs in Semnan [29]. Bacillus paralicheniformis like most of the above bacteria, has been isolated on various plants rather than the Figs. For example, from soybean paste [30], soil containing citrus waste soil in several regions of Golestan province [31], wheat fields of central provinces of Golestan and Mazandaran [27], rhizosphere of orange trees in Kerman province [28] and also from asparagus plant in Iran [29] and in New York [22,31].
Conclusion
In conclusion, it should be noted that all bacteria except Serratia marcescens are reported for the first time on Fig plants in Iran. There is no doubt that water, food and vegetables play an important role in the spread of pathogens and secondary infections. Given that some of these causal agents are transmitted through consumption of fresh fruits and vegetable, planting fruit trees in open gardens of hospitals is a major mistake and potentially very dangerous for the incidence and spread of diseases with a common human-plant hosts. And finally, due to the abundance of microbial populations in soft rot and damaged fruits, their consumption should be avoided to maintain the health of the body.
REFERENCES
- Jalili M. Small fruits. Urmia University Press, USA. (2nd edn). 2005;1-297.
- Sabetsarvestani J, Faghih H. Fig planting, having, harvest. Publishing House of Raqushah publishing house, Shiraz, Iran, 2001;1-292.
- Agricultural Statistics. Ministry of Jihad-e-Agriculture - Planning and Economic Deputy. Horticultural products (unfertilized and fertile levels, yields and yields broken down by water and rain). 2015;29-31.
- FAO. Statistical Database. 2012. Retrieved from htpp://faostat.Fao.Org.
- Palmateer AJ, Tarnowski TB, Roberts PD. Florida plant disease management guide: Fig (Ficus carica). IFAS, University of Florida. 2010;3-14.
- Anonymous. Fig diseases. 2017. Retrieved from Growables.org.
- Campoverde EV, Palmateer AJ. A severe outbreak of Xanthomonas on Ficus elasticin South Florida. Proc Fla State Hort Soc. 2011;124:321-322.
- Elboutahiri N, Thami-Alami I, Zaid E, Udupa SM. Genotypic characterization of indigenous Sinorhizobium meliloti and Rhizobium sullae by rep-PCR, RAPD and ARDRA analyses. Afr J Biotechnol. 2009;8(6):979-985.
- Bouzar H , Jones JB. Agrobacterium larrymoorei sp. nov., a pathogen isolated from aerial tumours of Ficus benjamina. Int J Syst Evol. 2001;51:1023-1026.
- Crisosto CH, Kader AA. The Commercial storage of fruits, vegetables, and florist and nursery stock. In: Gross KC, CY Wang, and M Saltveit (eds.) USDA, ARS, Agricultural Hdbk. 2004.
- Ferguson L, Erguson L, Bremer V , Colelli G. Fig (Ficus carica L.), pp. 134-158. In: Yahia Elhadi, M. (ed.) Postharvest biology and technology of tropical and subtropical fruits Vol. 3. Woodhead Publishing Limited, Cambridge. 2011;1-584.
- Henley D. An Overview of Ficus for interior landscapes. Greenhouse Manager. 1991;9(9):62-64.
- Caldis PD. Souring of gigs by yeasts and the transmission of the disease by insects. J Agri Res. 1930;40(11):1931-1951.
- Klement Z. Rapid detection of the pathogenicity of phytopathogenic Pseudomonas. Nature. 1963;199:299-300.
- Schaad NW. Laboratory Guide for Identification of Plant Pathogenic Bacteria. Second ed., API Press, 1988;1-158.
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A Laboratory Manual, (2nd edn) Cold Spring Harbor Laboratory Press, New York, USA. 1989;1-1656.
- Palacio-Bielsa A, Cambra MA, Lopez MM. PCR detection and identification of plant-pathogenic bacteria: updated review of protocols (1989-2007). J Plant Pathol. 2009;91:249-297.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30:2725-2729.
- Claus D, Berkeley RCW. Genus Bacillus Cohn, 1872. In: Sneath PHA, Mair NS, Sharpe ME and JG Holt, Edn, Bergey’s Manual of Systematic Bacteriology, The Williams & Wilkins Co., Baltimore, 1986;2:1105-1139.
- Aslani M, Hahsemipour M, Sadat NV, Shahcheraghi F, Eidi A, Sharafi Z. PCR identification of Pseudomonas aeruginosa based on two outer membrane lipoprotein oprI, oprL, and exotoxin A gene. Yafteh. 2009;11(2):21-26.
- Pereira JN, Morgan ME. Nutrition and physiology of Pseudomonas fragi. J Bacteriol. 1957;74(6):710-713.
- Edelman JR, Lin YJ. Microbiology (Endobacteriology) of fruit and vegetable crops: An expanded and continuing study. Int J Food Sci Nutr. 2016;5(2):95-104.
- Tahmasebi F, lakziyan A, Khavazi K, Pakdin PA. Isolation, characterization and evaluation of siderophore production in Pseudomonas bacteria and its effect on maize growth in aqueous media. J Cell Mol Res. 2014;27(1):75-87.
- Yu L, Qin XY, Du J, Wang AY, Zhao YY, Shen DJ, et al. Bacterial leaf spot of tobacco caused by Pseudomonas aeruginosa in China. Plant Pathol. 2008;57(4):774- 774.
- Habibi H, Khodaygan P. Identification of pathogenic bacteria with nucleus, grainy and walnut fruit trees in Alborz province. Msc Thesis. Valiasr University of Rafsanjan. 2013;83.
- Nagaraju V, Divakar G. Screening and isolation of pectinase producing bacteria from various regions in Bangalore. Int J Res Pharm Sci .2013;4(1):151-154.
- Etebarian H, Mohamadifar M, Alizade H, Zarisarabi A, Taheri P. Biological control of barley by antagonist bacteria. J Pest Plant Dis. 2008;74(82):81-91.
- Rezaei R, Alizadeh H, Azadvar M, Salari KH. Biocontrol properties of Bacillus species isolated from cranberry rosacea against Sclerotinia sclerotiorum. Biological Containment in Plant Protection. 2016;4(2):85-98.
- Moghbli M, Noshiri H. Isolation of a native strain of Bacillus licheniformisproducing alpha-amylase from warm sources of Semnan province. J World Micro. 2009;2(3):155-160.
- Christopher A, Dunlap Kwon SW, Alejandro PR, Soo-Jin K. Bacillus paralicheniformissp. nov., isolated from fermented soybean paste. Int J Syst Evol Microbiol. 2015;65:3487-3492.
- Mohammadi R, Dadgar T, Pardeli H, Yazdansetad S, Najafpour R, Farajtabrizi E. Molecular isolation and characterization of Bacillus cereus and Bacillus subtilisas pectinase enzyme producers from different regions of Golestan Province. J Cell Mol Res. 2016;29(3):340-348.
Citation: Mousavi SA, Hasanzadeh N, Abdollahi P (2020) Identification and Characterization of Bacterial Agents Causing Moderate Damage and Souring of the Fig Fruits. J Plant Pathol Microbiol. 11:514. doi: 10.35248/2157-7471.20.11.514
Copyright: © 2020 Mousavi SA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

