Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- CiteFactor
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2019) Volume 10, Issue 3
Genetic Variation among the Penicillia Isolated From Citrus under Storage Condition
Ahmad Ali Shahid, Mehra Azam, Muhammad Ali*, Kiran Nawaz and Moazzam AneesReceived: 21-Jan-2019 Published: 16-Feb-2019
Abstract
Pakistan is the sixth largest producer of Kinow (mandarin) and oranges in the world, with 2.1 million tons. In this research, fruit and vegetable markets in Lahore were visited and forty infected citrus samples were collected, surface sterilised and inoculated on potato dextrose agar medium for isolation. According to microscopy and colony characteristics five species of the genus Penicillium (P. digitatum, P. expansum, P. biourgeianum) were identified. Genomic DNA was isolated and internal transcribed spacer (ITS) was amplified by using primers ITS 1 and ITS4 and the 5.8S rRNA genes of fungi. Purified and amplified PCR products were later on sequenced and the sequenced homology was studied by BLAST and searching against databases of Genebank or NCBI software. The DNA sequence homology showed that isolates contain more than 98% homology with P. expansum, P. digitatum and P. biourgeianum known DNA sequences in the gene Data bank. Among these fungal isolates P. biourgieanum was another newly reported species isolated from infected citrus, and was assigned NCBI Gene bank accession number KT336317.
Keywords
Citrus; Molecular identification; Post-harvest pathology; White mold
Introduction
According to a report, Pakistan’s rank in citrus production is 10th worldwide. Furthermore, enormous economic losses happen all over the globe because of post-harvest diseases. According to a survey report it was concluded that 40% of all the citrus produced in Pakistan is lost in storage because of post-harvest diseases [1] and is commonly condensed by fungal pathogens attack [2]. Storage life as well as customer acceptability is minimized by the fungal infection during storage. Penicillium species are normally originate as contaminants in foods while drying and subsequent storage [3]. Identification of genus Penicillium is a difficult task because it is a large genus, and many common species look alike to the members of this genus. Moreover, there is a great variation within the same species which results in the inaccurate identification and thus need a comprehensive DNA based identification. Among these molecular based tools available, internal transcribed spacer ITS [4] and partial β-tublin gene sequences [5,6] have been used for identification of the Penicillium species. Penicillium italicum is responsible for the cause of blue mold disease in stored citrus and causes losses up to 25% in the worldwide production [7,8]. Once the fungi grow on citrus it excretes different types of mycotoxins [9]. Blue mold is one of the most economically important post-harvest diseases in the world. It is caused by Penicillium spp., amongst which P. expansum (Link) Thom is dominant. Blue mold decay disease produces patulin which is a carcinogenic mycotoxin and is also responsible for noteworthy economic losses throughout storage which effect the processing of stored fruits [10]. P. expansum contaminates fruit initially by means of wounds and these wounds are the caused during harvest or by the mishandling during storage process. The pathogen can also penetrate through natural openings, i.e., lenticels; stem ends and the calyx end [11]. Symptoms appear on the surface of fruit as light coloured soft lesions. Softness of the lesions is due to maceration of fruit tissues by the excretion of polygalacturonase enzyme which plays a noteworthy role in P. expansum virulence [12]. Fingerprinting methods generally used for identification purposes include Random Amplified Polymorphic DNA (RAPD). Amplified Fragment Length Polymorphic fingerprinting (AFLP). Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP) [13-15], micro arrays [16], DNA barcoding [17] and DNA sequencing [18]. Variations on PCR identification methods include multiplex PCR assays [19] and Real-Time PCR. Fungal identification can be rapidly performed as the ITS gene region [20] forms part of the highly conserved 5.8S ribosomal DNA. This region is present in most fungal species and commonly used primers are readily available.
Material and Methods
Isolation, identification and maintenance of pathogenic fungi
Different surveys were conducted to a number of fruit and vegetable markets (Badami Bagh, Lahore, Fruit and Vegetable market at Allama Iqbal Town, Lahore, Fruit and Vegetable market at Baghbanpura, Lahore and Fruit and Vegetable market at Ravi Link road, Lahore). Infected Apple samples were collected on the basis of their symptomatology under post-harvest storage conditions. Infected samples were surface sterilised and cut into 1cm small piece and inoculated on petri plates with PDA (Potato Dextrose Agar @ 37.0 g/L in distilled water) for isolation [21]. The inoculated plates were then incubated at 28°C for 6-7 days. After microscopy the fungal cultures were maintained on an agar slant at 4°C or stored as a few pieces of agar with mycelia in cryo vials with 10% glycerol in liquid nitrogen. Moreover, best suited media recipe for the growth and sporulation of pathogen was screened by growing inoculum on potato dextrose agar (PDA), malt extract agar (MEA) and corn meal agar (CMA). Inoculated plates were incubated at 28°C for 5-7 days.
Molecular characterization
Preparation of cultures for DNA isolation: The fungal colony was cultured in Potato Dextrose Broth (PDB) for 5-7 days at 28°C. The mycelial mat was harvested by filtering through a Whattsman filter paper and washed with sterile distilled water.
Genomic DNA isolation: 50 mg of mycelial mat was scrapped from 7 days old cultures. Culture mat was ground in pestle and mortar with liquid nitrogen. The powdered tissue was taken into a 1.5 mL micro-centrifuge tube. 500 μL CTAB buffer was added to the tube containing tissue and incubated for 1 h at 65°C. The supernatant was incubated with 10 μL RNase A solution (10 mg/mL) at 37°C for 1 h. after adding 500 μL chloroform and centrifuging at 13000 rpm for 5 min, samples were treated with isopropanol followed by washing the pellet with 70% ethanol to remove salts and let the pellet dry for 10 minutes. Finally pellet was resuspended in 50 μL of distilled H2O for further use.
Polymerase chain reaction amplification (PCR): The extracted DNA was subjected to the amplification with thermal cycler. PCR performed for the amplification of nuclear rDNA region containing the two Internal Transcribed Space r(ITS) regions (ITS1 TCCGTAGGTGAACCTGCGG and ITS4 TCCTCCGCTTATTGATATGC). The 44 μL of PCR master mixture containing 5 μL 10x Taq buffer, 5 μL 25 mM MgCl2, 2 μL 2 U/μL Taq polymerase enzyme, 5 μL 1 mM dNTPs, 5 μL 10 pmol ITS I, 5 μL 10 pmol primer ITS II, 17 μL Water H2O was used for each reaction: The 6 μL DNA template (10 μg/ μL) was added to the master mixture to make the final volume of the reaction 50 μL. The PCR reaction involved the initial denaturation at 94°C for 10 min followed by 30 cycles in series of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min each, with a final step of one cycle at 72°C for 10 min to final extension. The amplified products were confirmed at 1% agarose gel by using 100 bp DNA markers. The products were sequenced and identified on homology basis of their ITS regions, using BLAST at NCBI database.
Results and Discussion
Survey and sample collection
Four main fruit and vegetables markets in Lahore were surveyed and fruits were collected. Infected fruits were having fungal infection on their surface. A detailed map drawn to highlight the different collection sites is shown in Figure 1. The present research work was focussed mainly on the morphological and molecular identification of Penicillia, responsible for the loss of citrus fruits under storage conditions. During the survey of different fruit and vegetable markets (Figure 1) within Lahore it became evident that fruits are kept for more than a year under storage with different treatments so that, fruit supply can be ensure during whole year. It was also observed that mishandling during storage and improper storage place also contributes a lot towards the infection process.
Figure 1: A survey map showing sampling sites in Lahore, A; Fruit and vegetable market, Badami Bagh, B; Fruit and vegetable market, Allama Iqbal Town, C; Fruit and vegetable market, Baghbanpura, D; Fruit and vegetable market, Ravi link road.
Isolation and identification of pathogenic fungi
Penicillium biouregianum: Colonies were 15-25 mm in diameter, sparse, plane, surface texture voluminous to floccose; mycelium inconspicuous, margins wide, subsurface. Mycelium became greenish grey to greyish green or near globose due to heavy conidiogenesis. Reverse of the colony was pale, yellow to umber. Exudates and soluble pigment absent. Hyphae were septate and subsurface from which conidiophores arise with stipe commonly 200-400 μm in length. Stipe was smooth walled, it had metulae which were frequently apically inflated. Phialides were cylindrical. Conidia smooth walled, ellipsoidal to short cylindrical, 3-4 μm in diameter (Figure 2). Colonies were 15-20 mm in diameter, sparse, plane, with deeply floccose sectors; mycelium inconspicuous, margins wide, subsurface. Mycelium became greyer dull green due to heavy conidiogenesis. Reverse of the colony was pale to yellow. Conidiophores borne from subsurface or surface hyphae, with stipe commonly 200-350 μm long. Stipe was smooth walled, bearing terminal conidiophore, metulae and phialides were present and ampulliform to almost cylindrical. Conidia smooth walled, ellipsoidal, 3.0-3.5 × 2.5-3.0 μm (Figure 3). Colonies 35 -70 mm in diameter, plane, relatively sparse, very deep and floccose with an overlay of white mycelium, margins irregular. Mycelium became dull green due to moderate conidiogenesis. Reverse of the colony was pale or brownish (Figure 4A). Conidiophores borne from surface septate hyphae. Stipe was smooth walled, metulae and Phialides were ampulliform to cylindrical. Conidia smooth walled, ellipsoidal to cylindroidal, 6.0-8.0(-15) × 2.5-5.0-(6.0) μm (Figure 4B). Ivana et al., [22] have concluded similar findings during characterization of molds from apples under storage. Infected tissues were seen to be macerated. Three isolates of the genus Penicillium were identified by their morphological characterization. These isolates had variation in their colony shape, color, size, and other microscopic traits such as hyphal septation, conidia and metulae. Raphael et al., [23] also discussed the same macroscopic as well as microscopic features of P. expansum while working with the biological control of these two pathogenic fungi infecting citrus fruits in Brazil.
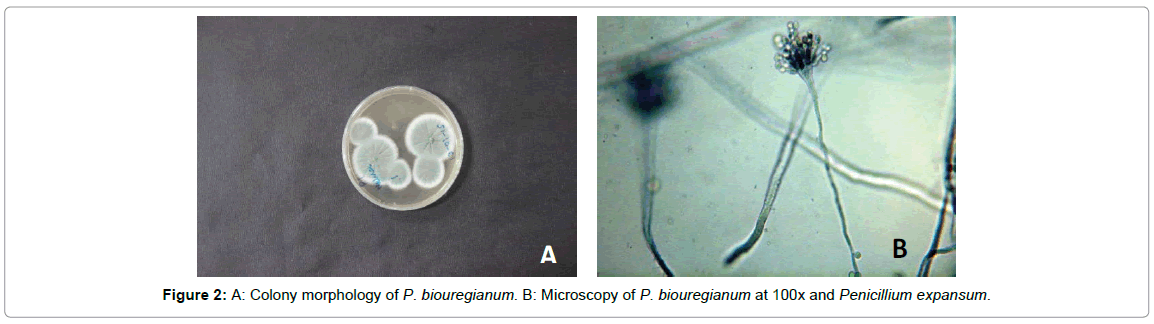
Figure 2: A: Colony morphology of P. biouregianum. B: Microscopy of P. biouregianum at 100x and Penicillium expansum.
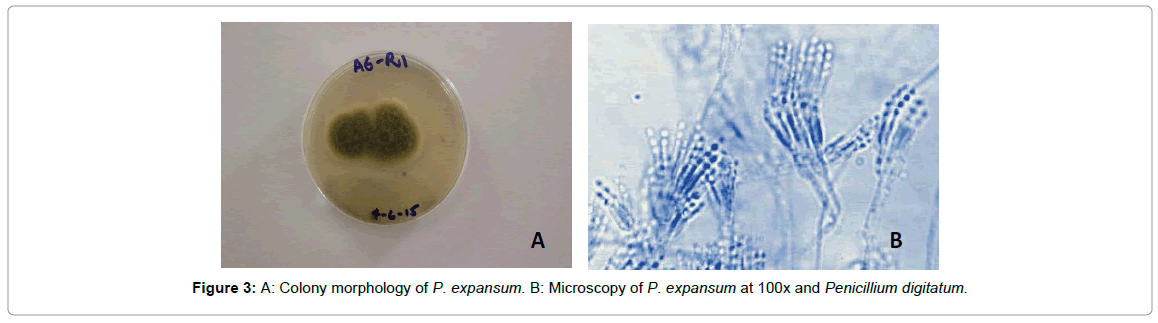
Figure 3: A: Colony morphology of P. expansum. B: Microscopy of P. expansum at 100x and Penicillium digitatum.
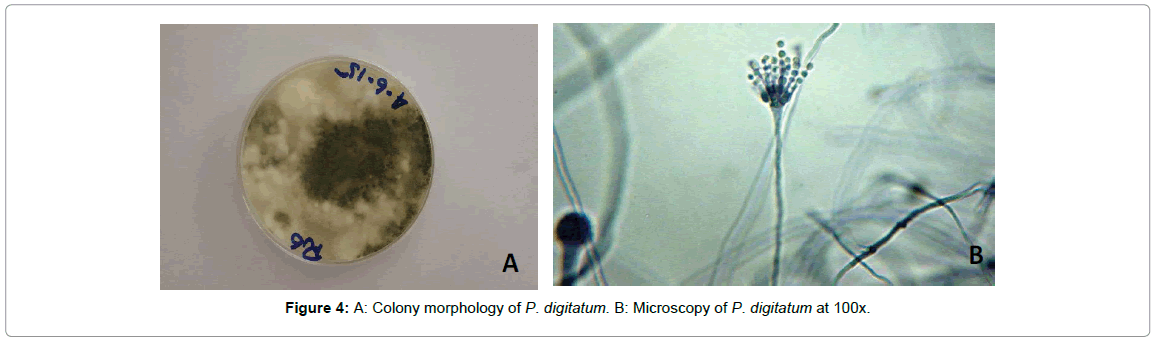
Figure 4: A: Colony morphology of P. digitatum. B: Microscopy of P. digitatum at 100x.
Genomic DNA extraction: Genomic DNA was extracted of all 8 selected Penicillium isolates. Extracted DNA was checked by gel electrophoresis under UV light in gel documentation system and picture was captured using GRAB IT programme. For the amplification of nuclear rDNA region containing the two Internal Transcribed Spacer (ITS) regions (ITS1 and ITS4) and the 5.8S rRNA gene in the Penicillium and Aspergillus sp. ITS PCR was performed with primer pair ITS1/4 at annealing temperature of 55°C. The length of the ITS region alignment used was 330-1500 bp. In this study, Genomic DNA was extracted and quantified in the range of 330 to 1500 bp and photographed by using GRAB IT programme. Gel Picture shows a good quality of extracted DNA. All genomic DNA size was range from 330 to 1500 bp nucleotides. For the amplification of nuclear rDNA region containing the two Internal Transcribed Spacer (ITS) regions (ITS1 and ITS4) and the 5.8S rRNA gene in the Penicillium. ITS PCR was performed with primer pair ITS1/4 at annealing temperature of 55°C. The length of the ITS region alignment used was 330-1500 bp (Figures 5 and 6). These results are in conformity with the findings of White et al.
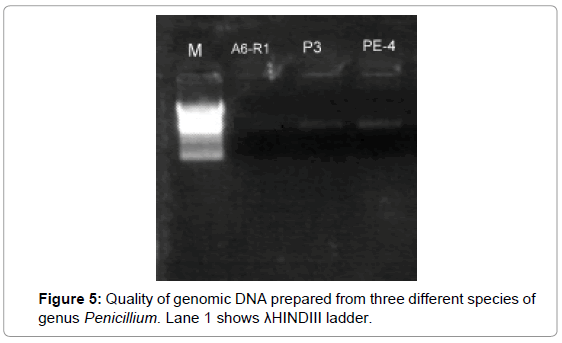
Figure 5: Quality of genomic DNA prepared from three different species of genus Penicillium. Lane 1 shows λHINDIII ladder.
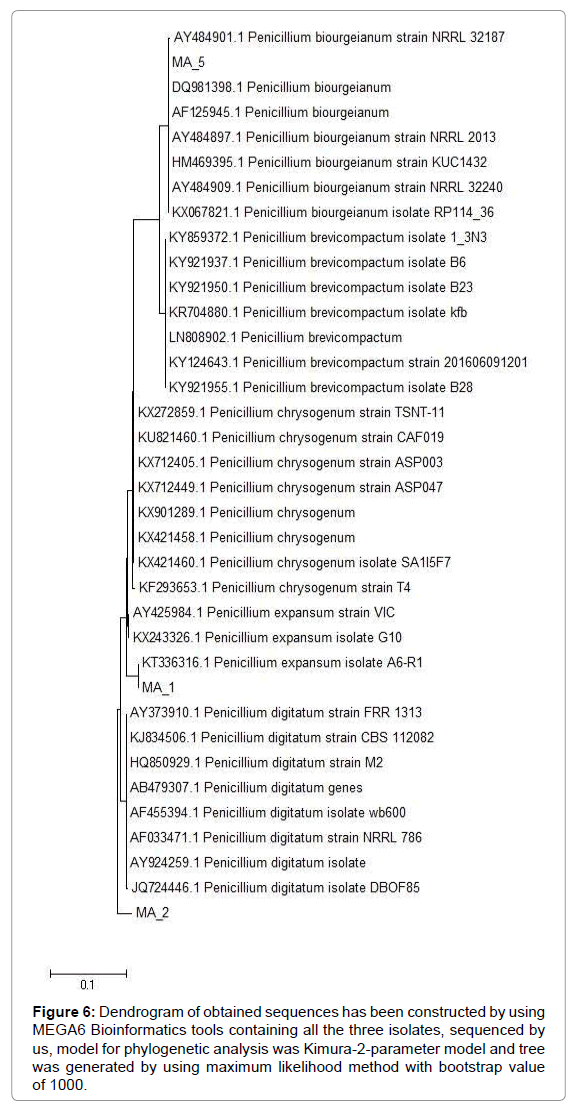
Figure 6: Dendrogram of obtained sequences has been constructed by using MEGA6 Bioinformatics tools containing all the three isolates, sequenced by us, model for phylogenetic analysis was Kimura-2-parameter model and tree was generated by using maximum likelihood method with bootstrap value of 1000.
Phylogenetic identification: Phylogenetic dendograms were made by using maximum likelihood method in Mega6 software, on the basis of most relevant sequences obtained by blasting my sequence on NCBI. Two Penicillia (P. expansum and P. biourgieanum) were first time reported from Pakistan, submitted to NCBI with accession numbers KT336315 and KT336317 respectively. Phylogenetic analysis of the isolated Penicillium has revealed that P. biourgieanum was closely related to the isolates of P. biourgieanum (Accession No. AY484901) from Peoria, USA isolated and characterized by Peterson, 2004 and P. digitatum have shown maximum likelihood with P. digitatum (Accession No. 724446) from Tamil Nadu, India. Isolated and characterised by Kumar, 2013. In the same way P. expansum have exhibited maximum homology with P. expansum (Accession No. KX243326) from USA, isolated by Yin (Table 1).
| Serial No. | Accession No. | Organism |
|---|---|---|
| 1 | KT336316 | Penicillium expansum |
| 2 | KT336317 | P. biouregieanum |
Table 1: NCBI accession numbers assigned to pathogens.
Conclusion
In a nutshell it is evident from the above discussion that the genus Penicillium some other fungal isolates were also studied during isolation. But most abundant were the species of P. italicum, P. expansum and P. biouregieanum. These fungal species are actually responsible for the green mold, fruit rot, spoilage and other loss of citrus fruit in storage conditions. Level of damage to citrus fruit under storage conditions due to different fungal isolates was also studied. Phylogenetic analysis of dendrogram has shown similarity within the same species as well as among different species of genus Penicillium by which actual genetic variability among the genus penicillium was seen.
REFERENCES
- Naseer M (2010) Government of Pakistan, A report on export of Trade Development Authority of Pakistan, Faisalabad, Pakistan.
- Liu Q, Xu J, Liu Y, Zhao XL, Deng XX, et al. (2007). A novel bud mutation that confers abnormal patterns of lycopene accumulation in sweet orange fruit (citrus sinensis l. Osbeck). J Exp Bot 58: 416-417.
- Harman GE, Howell CR, Viterbo A, Chet I, Lorito M (2004) Trichoderma species opportunistic, avirulent plant symbionts. Nature Reviews Microbiology 2: 43-56.
- Peterson SW (2004) Multilocus DNA sequence analysis shows that Penicillium biourgeianum is a distinct species closely related to P. brevicompactum and P. olsonii. Mycolo Res 108: 434-440.
- Seifert KA, Louis-seize G (2000) Phylogeny and species concepts in the Penicillium aurantiogriseum complex as inferred from partial ~-tubulin gene dna sequences. In: integration of modern taxonomic methods for Penicillium and Aspergillus classification. Samson, R.A. and Pitt, J.I. (eds.) pp: 189-198.
- Samson RA, Seifert KA, Kuijpers AFA, Houbraken JAMP, Frisvad JC (2004) Phylogenetic analysis of Penicillium subgenus Penicillium using partial p-tubulin sequences. Studies in mycology 49: 175-200.
- Palou LA, Marcilla C, Rojas-argudo M, Alonso JA, Del rio MA (2007) Effects of x-ray irradiation and sodium carbonate treatments on postharvest Penicillium decay and quality attributes of clementine mandarins. Postharvest Boil Tec 46: 256-261.
- Montesinos-Herrero C, Del Rio MA, Paster C, Brunetti O, Palou L (2009) Evaluation of brief potassium sorbate dips to control postharvest Penicillium decay on major citrus species and cultivars. Postharvest boil Tec 52: 117-125.
- Moss MO (2008) Fungi, quality and safety issues in fresh fruits and vegetables. J Appl Microbiol 104: 1239-1243.
- Barkai-golan R (2008) Penicillium mycotoxins. In r. Barkai-golan & paster, n. (eds) mycotoxins in fruits and vegetables.pp:153-185.
- Rosenberger DA, Engle CA, Meyer FW, Watkins CB (2006) Penicillium expansum invades apples through stems during controlled atmosphere storage. Online. Plant health progr.
- Jurick WM, Vico I, Gaskins VL, Garrett WL, Whitaker BD, et al. (2010) Purification and biochemical characterization of polygalacturonase produced by Penicillium expansum during postharvest decay of ‘anjou’ pear. Phytopatholol 100: 42-48.
- Sequerra J, Marmeisse R, Valla G, Normand P, Capellano A, et al. (1997) Taxonomic position and intraspecific variability of the nodule forming Penicillium nodositatum inferred from RFLP analysis of the ribosomal intergenic spacer and random amplified polymorphic DNA. Mycologi Rese 101: 465-472.
- Colombo A, Drzewiecki J, Banning A, Grube E, Hauptmann K, et al. (2003) Randomized study to assess the effectiveness of slow-and moderate-release polymer-based paclitaxel-eluting stents for coronary artery lesions. Circula 108: 788-794.
- Latouche GN, Huynh M, Sorrell TC, Meyer W (2003) PCR-restriction fragment length polymorphism analysis of the phospholipase B (PLB1) gene for subtyping of Cryptococcus neoformans isolates. Appl Environmen Microbiol 69: 2080-2086.
- Schmidt-Heydt M, Geisen R (2007) A microarray for monitoring the production of mycotoxins in food. Int J Food Microbiol 117: 131-140.
- Seifert JR, Mlodzik M (2007) Frizzled/PCP signalling: A conserved mechanism regulating cell polarity and directed motility. Nat Revi Geneti 8: 126.
- Vogler EA (1998) Structure and reactivity of water at biomaterial surfaces. Adv Colloid Interfa Sci 74: 69-117.
- Dean RA, Talbot NJ, Ebbole DJ, Farman ML (2005) The genome sequence of the rice blast fungus Magnaporthe grisea. Natu 434: 980.
- White TJ, Bruns T, Lee SJWT, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications 18: 315-322.
- Gutierrez CG, Glazyrin AL, Robertson GW, Campbell BK, Gong JG, et al. (1997) Ultra-structural characteristics of bovine granulosa cells associated with maintenance of oestradiol production in vitro. Mol Cellu Endocrinol 134: 51-58.
- Ivana V, Nataša D, Miljan V, Milica N (2014) Identification of Penicillium expansum causing post-harvest blue mold decay of apple fruit. Pestic Phytomed (belgrade) 29: 257-266.
- Raphael S, Pimenta L, Francisco L, Silva J, Silva F, et al. (2008) Biological control of Penicillium italicum, P. digitatum and P. expansum by the predacious yeast Saccharomycopsis schoenii on oranges. Braz J microbio 39: 85-90.
Citation: Shahid AA, Azam M, Ali M, Nawaz K, Anees M (2019) Genetic Variation among the Penicillia Isolated From Citrus under Storage Condition. J Plant Pathol Microbiol 10:474.
Copyright: © 2019 Shahid AA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

