Indexed In
- Academic Journals Database
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- China National Knowledge Infrastructure (CNKI)
- Scimago
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- University Grants Commission
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Review Article - (2023) Volume 15, Issue 3
Exploring the Potential of Genetically Engineered Whole-Cell Biosensors: A Step towards Device Development for Environmental Monitoring
Nasreen S Munshi*, Hinal Mehta, Aarushi Sachdeva and Jinal BhavsarReceived: 30-Jun-2023, Manuscript No. JMBT-23-21974; Editor assigned: 03-Jul-2023, Pre QC No. JMBT-23-21974 (PQ); Reviewed: 17-Jul-2023, QC No. JMBT-23-21974; Revised: 24-Jul-2023, Manuscript No. JMBT-23-21974 (R); Published: 31-Jul-2023, DOI: 10.35248/1948-5948.23.15.558
Abstract
Monitoring and detection of hazardous contaminants present in wastewater are mandatory for wastewater management at precise stages. Hence, the biosensor is one such detection device for real-time monitoring of contaminant levels in wastewater with user-friendly technology. A key innovation in the field of bio sensing occurred through genetically engineered whole- cell biosensors. Here, the cells are genetically engineered by introducing reporter genes and expressions of those genes are controlled by promoter sequences/regulatory proteins. They are modified in a way to achieve analyte specificity based on their sensing capabilities and coupling them to a transducer. Advancements in accessible biosensors are not only reliant on genetic engineering but also on the methods for their biocompatible immobilization and fabrication. To minimize the number of handling steps in the future for field application, it is feasible to immobilize cells by trapping them inside a polymer matrix, keeping them in the exceptional chambers of microfluidic devices, or simply allowing them to adhere to the paper. In this review, the emphasis is on the biosensing strains, immobilization techniques, multiple matrices on which cells can be immobilized, as well as the strategies for device development. Several biosensing strains have been described concerning their reporter genes targeting different contaminants present in wastewater. This review is intended to provide interesting information, especially on the strategy planned to immobilize biosensing strains while retaining their long-term viability and reproducibility for function. It is anticipated that it provides insights into a systematic and fundamental understanding of fabricating the efficient transduction system, as well as inspire for development of smart devices in nearby future for environmental monitoring.
Keywords
Contaminants; Pollutants; Whole-cell biosensors; Immobilization; Matrices; Biosensor device
Abbreviations
FP:Fluorescent Protein; PAH:Polycyclic Aromatic Hydrocarbon; YFP:Yellow Fluorescent Protein; RFP:Red Fluorescent Protein; APS:Ammonium Persulphate; SDW:Synthetic Dye Wastewater; PVA:Polyvinyl Alcohol; PAMs:Polyacrylamides; GFP:Green Fluorescent Protein; DGT:Diffusive Gradients in Thin Films Technique; PDMS:Polydimethylsiloxane; CCD:Charge Coupled Device Lab; VIEW:Laboratory Virtual Instrument Engineering Workbench; PMT:Photomultiplier; LLG:Liquid Light Guide
Introduction
Massive urbanization with its improved living standards and expansion in industrialization along with various anthropogenic activities are introducing multitudinous hazardous contaminants directly to the environment [1]. These contaminants such as heavy metals, polycyclic hydrocarbons, and other inorganic pollutants released into various water bodies are mostly in a concentration toxic enough to cause irreversible effects on environment and human health [2,3]. Monitoring and detection of such contaminants in water bodies as well as wastewater are mandatory for environmental management at precise stages [4]. Therefore, the biosensor is one such detection device for real-time monitoring of contaminant levels in wastewater with user-friendly technology [5,6]. The biosensor is a device comprising of biomolecule recognition elements (such as whole-cell, enzyme-based, antigen-antibody based, and nucleic-acid based) that is in direct contact with the target molecule and in close proximity to transducer (electrochemical, optical, piezoelectric and thermal) converting a recognition signal into measurable output [4,7-10]. A key innovation in biosensing occurred through microbial biosensing strains. Researchers have designed various biosensing strains having a fusion of reporter gene to promoter- operator sequence of hydrocarbon-degrading gene/operon [11]. There are different types of frequently used reporter genes which can be used in a biosensor, e.g. lux based reporter genes produce light signals, GFP gene based reporter gene (and related genes like RFP, CFP, YFP, etc.) produces green fluorescent protein signals (or colour equivalent to reporter gene), enzyme based reporter gene produces catalytic signals identified by either production of the product or depletion of substrate of the enzyme.
A genetically modified whole-cell biosensor specific for any contaminant, having Fluorescent Protein (FP) based reporter system produces fluorescent proteins that will fluoresce in the presence of that contaminant [12]. The fluorescence emitted from fluorescent protein can be detected qualitatively as well as quantitatively through an electronic system as shown in Figure 1. An appropriate biosensing strain is the one which is designed in a way to generate signals which assist to measure the level of contaminants. This process will also emphasize on an important aspect of contaminant, i.e. its bioavailability. For successful bioremediation of any organic pollutant, its bioavailability should be high.
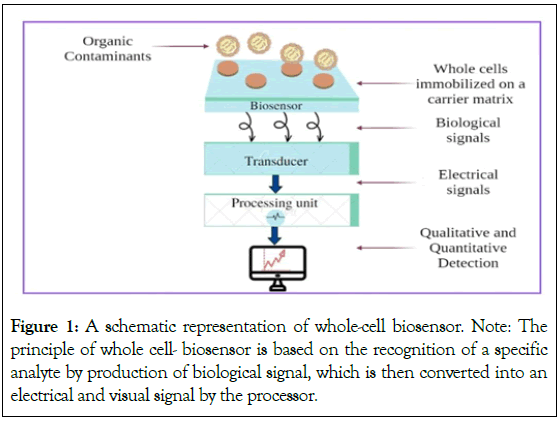
Figure 1: A schematic representation of whole-cell biosensor.
Note: The
principle of whole cell- biosensor is based on the recognition of a specific
analyte by production of biological signal, which is then converted into an
electrical and visual signal by the processor.
Various ranges of reporter genes are available (such as lux (bacteria), luc (firefly), GFP (jellyfish-Aequorea victoria and its variants), and lacZ (E. coli)) which can be fused with different promoter-operator systems [13]. This fusion can be advantageous to detect multiple contaminants simultaneously through multiple biosensing strains generating a range of diverse colors [14]. Whole-cell biosensors are proven to be favourable because of the speedy detection of contaminants, cost-effectiveness, and high-sensitivity in monitoring as compared to analytical methods [15]. Some of the advantages along with challenges and capabilities are mentioned in Figure 2.
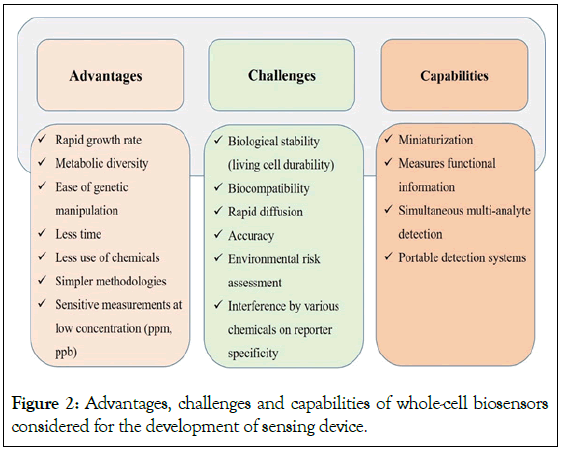
Figure 2: Advantages, challenges and capabilities of whole-cell biosensors considered for the development of sensing device.
Apart from the benefits of whole cell biosensors, it has limitations such as the multiplication of biosensing strains in the sensing system which may cause discrepancies in detection of fluorescence as depicted in Figure 2. In order to avoid this situation, biosensing strains can be immobilized which might improve the detection process as well as stabilize the cells [1]. Whole-cell biosensors have physical units on which cells can be immobilized. A reaction taking place on the surface of the transducer by the biosensing strains to bring out an electrical signal is the primary concept behind the biosensing system. In this progressing era of biosensing technology, the most sensitive footstep is to immobilize bio recognition elements (biosensing strains) onto transducers. The selection of appropriate immobilization techniques may contribute to the development of an efficient, precise, and cost-effective biosensor that can be commercialized undoubtedly. The biosensing strains should remain alive on the matrix in order to execute their sensing activities. Hence, any of the mechanisms selected for cell fixation such as immobilization, encapsulation, or adherence, must withstand their viability and functionality [16-18]. The most common polymer/gel- based methods available for the immobilization of biosensing strains are agar, agarose, alginate, polyacrylamide, gelatin, and chitosan. These are natural matrices that do not affect the growth and activity of microorganisms. Another well-accomplished technique is soft lithography which includes the association of polymers with microfabrication (which is a technique for making items with dimensions between a few micrometers to a few millimeters using integrated circuit manufacturing technology) for the integration of biosensing strains onto biosensor platforms [19].
The behaviour of biosensing strains is required to be monitored by various transduction systems (optical, electrochemical, optoelectronic etc.), where the signal is generated due to the genetic response of the cells. Integration of viable biosensing strains with microelectronic technologies requires a scientific solution as well as optimized signal processing technique [20]. The resultant signal from the cell’s response will be stored and analysed. Researchers have successfully demonstrated the integration of biosensing strains into microfluidic systems which could directly display the level of the contaminants through fluorescence signals. This sort of visual assessment would be easy for retrieving data and interpretation [17].
Construction of Biosensing Strains
Recombinant DNA technology has helped researchers to engineer microbes. They started using molecular techniques to house foreign genes of interest that would produce a quantifiable signal in response to a target analyte. In normal bacterial cells, the promoter is linked to other genes which are transcribed and then translated into proteins helping the cell to adapt and utilize the exposed analyte as a carbon source [14]. Whereas, biosensing strains contain two genetic elements: promoter and reporter. When analyte comes in contact with cell, it can interact with it in two ways as shown in Figure 3. A: When detection of analyte requires only one regulatory protein and promoter which can sense the pollutant and transmit the signal to its reporter gene, which ultimately produces signaling molecule; B: In second case, where analyte first binds to the receptor present on the cell surface and provides the signal to the regulatory gene. Upon sensing of pollutant by regulatory gene, it induces the promoter and finally activates the reporter gene leading to production of detectable signaling molecules [14]. Symbol key represents the cellular entities of the cell. In these strains, the gene situated downstream is expressed to produce a reporter protein that generates a detectable signal when the promoter is activated by the pollutant acting as an inducer molecule. The promoter sequence is generally derived from an operon which is involved in the degradation of an organic pollutant. Thus, the biosensing cells have a plasmid with a fused promoter-operator and a reporter gene that function as a sensing circuit.
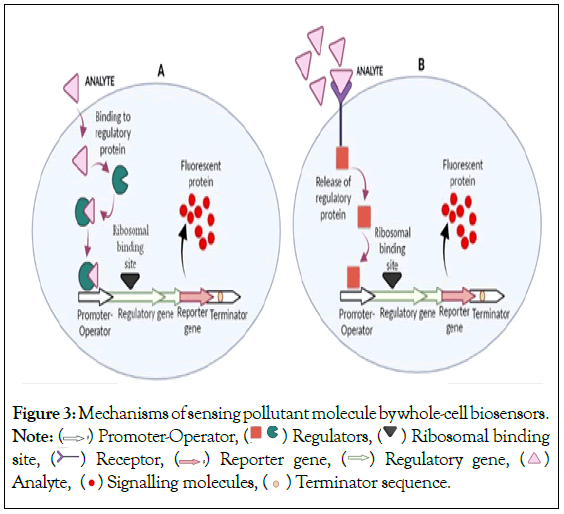
Figure 3: Mechanisms of sensing pollutant molecule by whole-cell biosensors.
Note:  Promoter-Operator,
Promoter-Operator,  Regulators,
Regulators,  Ribosomal binding site,
Ribosomal binding site,  Receptor,
Receptor,  Reporter gene,
Reporter gene,  Regulatory gene,
Regulatory gene,  Analyte,
Analyte,  Terminator sequence.
Terminator sequence.
To date, many biosensing strains have been constructed with reporter gene LuxAB. Despite being one of the most common reporter genes, LuxAB emits only a single type of signal which is in the form of bioluminescence, making it difficult for researchers to differentiate between two or more types of contaminants [14]. Whereas, using a diverse range of Fluorescent Protein (FP) genes available nowadays, numerous promoter-operator systems with distinct FP genes could be tagged to work under natural environments. Multiple contaminants can be determined simultaneously, with diverse colour signals emitted by a variety of FP integrated as reporter genes [21]. Fluorescence-based biosensors have their own fixed excitation and emission wavelengths [22].
In 1975, Divies created the first microbial biosensor (Acetobacter xylinum) for ethanol detection [23]. This initiative became the base for the development of microbial sensors for monitoring purposes. The alk regulon was used for the detection of alkanes, the nahG gene of a naphthalene catabolic plasmid in Pseudomonas fluorescent was used for the detection of naphthalene, and the TOL system was explored for the detection of benzene derivatives, to name a few bioluminescent strains which were used for the detection of hydrocarbons constructed a biosensor named RP007 strain of Burkholderia sartisoli having the construct probe-phn-luxAB for the detection of aromatic hydrocarbons such as naphthalene and phenanthrene in seawater [24-26]. The modified strain was able to detect hydrocarbon in seawater after 3 h of incubation with a lower detection limit of 0.17 µM. Here, the biosensing strains were constructed by cloning the hydrocarbon sensing promoter-operator region which was fused with luxAB as reporter genes. Patel et al. developed a novel strategy in which two biosensing strains were tagged with two specific types of FP reporter genes for the detection of different categories of aromatic hydrocarbons differentially [14]. Initially both the strains had luxAB as the reporter gene. For the detection and quantification of mono- and poly-aromatic hydrocarbons, two clones named as E. coli 2296 and E. coli 2301 were created by using a fusion of promoter-operator and reporter genes such as tbut-gfp and phnR-cfp respectively. The promoter-operator tbuT in E. coli 2296 is capable of detecting BTEX compounds and promoter-operator phnR in E. coli 2301 is capable of detecting naphthalene, phenanthrene, and other PAH (Polycyclic Aromatic Hydrocarbon) compounds. The designed vectors were transformed into E. coli DH5α, and these strains were designated as E. coli DH5α 2296-gfp (containing pPROBE-tbuT- RBS-gfp-npt) and E. coli DH5α 2301-cfp (containing pPROBE- phn-RBS-cfp-npt). Both the recombinant strains were capable of detecting hydrocarbons in the range of 1-100 µM range.
Hu et al. constructed a yellow fluorescent protein (phiYFP) based whole-cell biosensor for the detection of arsenite and arsenate [27]. An arsenic-resistant promoter and regulatory gene arsR were obtained by PCR (Polymerase Chain Reaction) from E. coli DH5α. phiYFP was inserted as a reporter gene in arsenic resistant Whole- Cell Biosensor (WCB-11) for its expression. The constructed whole- cell biosensor showed a good response when exposed to different concentrations of arsenic.
Similarly, a whole-cell biosensor was constructed for the detection of mercury pollutants in cosmetics such as mercuric chloride (HgCl2), Mercurous chloride (Hg2Cl2), and mercuric ammonium chloride (Hg(NH2)Cl) by Guo, et al. [28]. The genetic circuits for the biosensor constitutively synthesized merR as sensor proteins and inducible rfp (red fluorescent protein) served as a reporter protein. The fluorescence intensity of rfp in this biosensor relatively showed a linear relationship (R2=0.9848) with the Hg (II) concentration ranging from 50 nM to 10 αM. Looking to the metal concentrations detected, biological sensors based on whole- cell systems seem to be highly valuable and useful.
A new microbial biosensor for heavy metal detection was constructed by Kim, et al [29]. by using cadC-cadO as a regulator- operator sequence obtained from Bacillus oceanisediminis 269. This strain was used because it had an operon that provided high level of resistance to heavy metal. Yet another novel and highly specific recombinant biosensor were constructed for the detection of highly toxic cadmium (Cd (II)) in milk samples by using E. coli DH5α pNV12. The reporter gene, gfp was introduced that was under the control of the cad promoter and cadC regulatory gene of Staphylococcus aureus plasmid pI258. The response time of the constructed biosensor was as low as 15 min. The linear range for detection of cadmium ion concentrations was 10 µg/l-50 µg/l with R2 of 0.9946 with a lower detection limit of 5 µg/l [30].
Immobilization of Whole-Cell Biosensors
The localization or physical confinement of entire cells to a specific area of surface without sacrificing desirable biological activity is referred to as the immobilization of whole cells [31]. Immobilization gives the microorganism a particular stability against environmental disturbances and shields the cells from shear pressures such as pH, organic solvents, temperature, salts, etc. Immobilized cells may be kept active, viable, and productive for an extended period of time [18]. Moreover, it enhances the biological stability of the cells [32,33]. Utilizing artificial or natural polymers, cell immobilization enables the effective restriction of cell movement, allowing the employment of free cells in place of biocatalysts. Cells can be immobilized by adhering to or becoming trapped in organic or inorganic, water-insoluble substances [34].
In comparison to enzyme immobilization, immobilization of whole cells seems to be more feasible, since it allows for efficient catalyst recovery, solids and liquid separation, and purification of the mixture [35]. The kind of application as well as the physical and biological properties of the immobilizing matrix or agent determines the usefulness of a particular method of immobilization [36]. In an immobilization procedure, the choice of carrier or matrix is crucial since it affects the viability of immobilized microorganisms. The support matrix or carrier used to immobilize cells must be biocompatible. There are several techniques for immobilization of microbes which are mentioned in Figure 4.
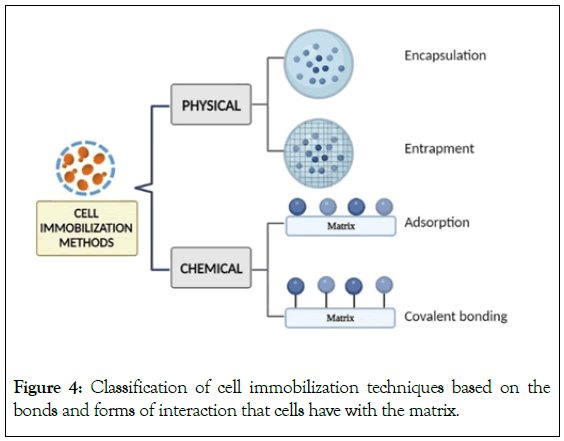
Figure 4: Classification of cell immobilization techniques based on the bonds and forms of interaction that cells have with the matrix.
Encapsulation
It is a physicochemical process of immobilization [37]. Encapsulation is an irreversible immobilization technique. It is a kind of entrapment in which the biocatalyst (cell) is contained within a semipermeable membrane where immobilized cells are free to float within the core space as depicted in Figure 5. While the cells are constrained by the membrane walls, the semi-permeability of the membrane permits the movement of substrates and nutrients through it. This technique restricts access to the capsules inside, protecting the cells from the outside environment and preventing leakage [34].
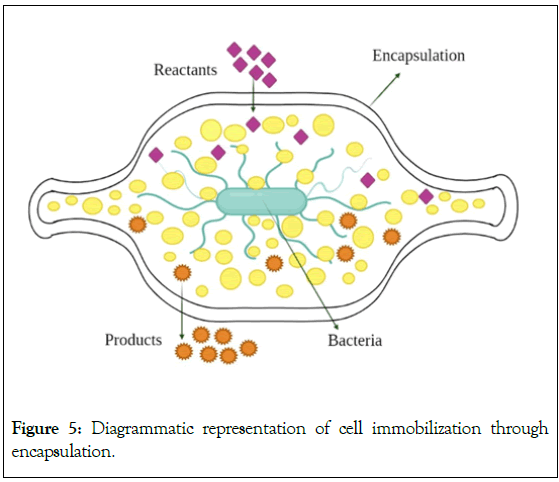
Figure 5: Diagrammatic representation of cell immobilization through encapsulation.
Entrapment
Entrapment is one of the frequently used techniques of cell immobilization. Microorganisms are trapped in a stiff network using entrapment techniques to restrict cell escape into the surrounding environment [38]. It is also an irreversible technique like encapsulation [39,40]. Compared to surface immobilization such as that occurs in adsorption, entrapped cells may attain great density in the matrix and are shielded from fluid shear forces [31]. The price of immobilization, diffusion restrictions, deactivation during immobilization, and breakage of support material during use are some drawbacks associated with this form of immobilization [40].
The support materials/matrices used for entrapment can be divided into thermo gels (cellulose, agarose, agar, etc.), hydrogels (alginate, chitosan, etc.) and synthetic polymers (polyurethane, Polyvinyl Alcohol (PVA), etc.) [38,41]. Although natural polymers are non-toxic, but they mostly have low durability and poor mechanical strength. On the contrary, the synthetic polymers have good mechanical strength, are available at economical rates and are preferable for immobilization. However, they can be toxic to microbes [41]. Entrapment in sodium alginate is one of the most popular and easy techniques. After combining with entire cells, the water-soluble alginate is dropped into a calcium chloride solution as depicted in Figure 6 to generate water-insoluble alginate beads [38]. This technique maintains the viability of cells in alginate gel. For application in soil conditions, a bacterial consortium comprising of seven isolates of acid tar-contaminated soil was immobilization in alginate gel. The purpose of this study was to find the right conditions for entrapping a novel bacterial consortium of Acinetobacter sp., Bacillus circulans, Bacillus licheniformis, Brevibacillus brevis, Burkholderia cepacia, Leifsonia aquatic, and Sphingomonas paucimobilis suited for the bioremediation of oil-contaminated soils [42].
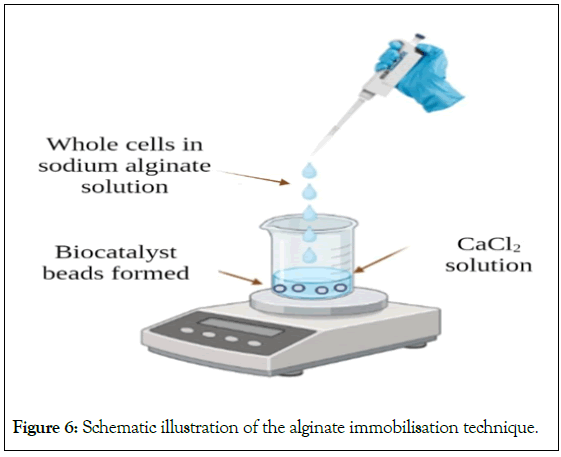
Figure 6: Schematic illustration of the alginate immobilisation technique.
One of the immobilization methods based on synthetic polymers uses polyacrylamide gel and it is another extensively utilized technique. Whole cells, acrylamide (monomer) and bis-acrylamide (a cross- linker reagent) are combined and polymerized with the help of an initiator like APS (Ammonium persulphate). Also, a polymerization accelerator like N, N, N’, N’-tetramethylethylenediamine is used which accelerates or enhances the polymerization [38].
Other matrices that are extensively used are agar and agarose [38]. For example, a bacterial consortium of Enterobacter dissolvens AGYP1 and Pseudomonas aeruginosa AGYP2 was immobilized on agar. The immobilized bacterial consortium was used for the effective decolourization of textile dyes from the Synthetic Dye Wastewater (SDW). When the consortium was immobilized on agar, 96% of the dye (100 mg/l) was removed within 6 h [43].
Adsorption
In adsorption, there is physical attachment of the microbe to the surface of a carrier that is water insoluble. It is one of the most widely used immobilization methods because of its reversible nature. When the cells are immobilized through adsorption, the cells are in direct contact with the nutrients. The transfer of the immobilized cells from the bulk to the exterior of the support is the first step in the cell immobilization by the adsorption process [34]. The interaction between the cell and the carrier is through ionic interactions, Van der Waals forces and/or hydrogen bonding. These molecular forces are weak and aid in prevention of change in structure of the immobilized cells [44]. Adsorption-based immobilization is mild, rapid, easy, cost-effective, and doesn’t require any chemical agents. Additionally, the procedure is simple to carry out and offers the option of reloading the support. This approach has some drawbacks because of unstable bonds and the inability to adjust the loading. This disadvantage might be able to cause the high rate of matrix leakage. Furthermore, the reproducibility is also low [45].
Covalent bonding
In this kind of immobilization, a covalent bond is formed between the activated support matrix and microbial cell [36,39]. Chemical surface modification is required for covalent bonding. Although covalent attachments are efficient and long-lasting for enzymes, they are rarely used to immobilize cells. The primary reason may be the fact that covalent bonding agents are frequently cytotoxic [46]. It is a cost-effective immobilization method. Covalent bonds connect the functional groups on cells to the carrier matrix. Contrary to adsorption, covalent bonding is an irreversible technique [34,47].
Carrier materials for cell immobilization
The choice of the matrix is crucial in the immobilization process because it affects the survival of the immobilized microorganisms and, consequently, the effectiveness of the biosensing system that will use the immobilized cells. The microbial cell support matrix needs to be insoluble, incapable of being broken down by biomass, non-toxic, affordable, and simple to handle. Additionally, it must have good mechanical, chemical and optical properties as well as biological stability. It must allow the optimal nutrient diffusion [34]. There are three different types of carriers as mentioned in Figure 7, viz. organic, inorganic and composite carriers [34]. Different kinds of matrices can be used to immobilize cells. Inorganic matrices are widely used because of their thermo stability as well as their resistance to microbial degradation. Several synthetic matrices can be used like polyurethane, polyvinyl alcohol, acrylamide and resins. However, many natural polymers can be used for the immobilization of cells like algal polymers (carrageenan, alginate, agar and agarose) and chitosan which is a fungal derivative [39]. The most popular method for immobilizing live cells is to trap them inside spheres of Ca2+ alginate [48-50]. The frequently used matrices for the development of biosensors with whole-cells immobilized are described here.
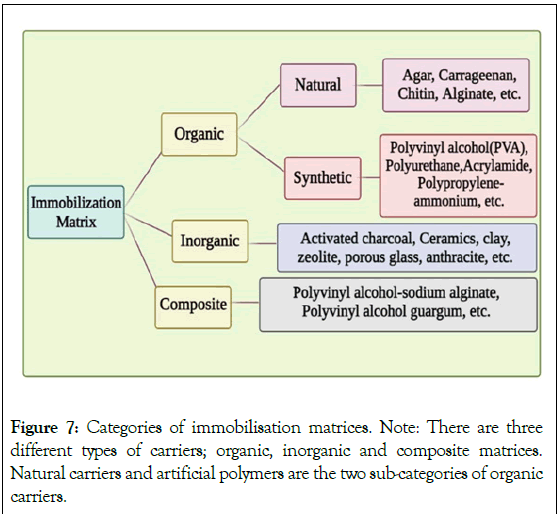
Figure 7: Categories of immobilisation matrices.
Note: There are three different types of carriers; organic, inorganic and composite matrices.
Natural carriers and artificial polymers are the two sub-categories of organic
carriers.
Alginate
Alginates or alginic acids are natural polysaccharides. These are a class of anionic unbranched polysaccharides which are derived from the cell walls of the brown algae/seaweeds (Phaeophycea), which are found in the shallow waters of temperate regions [51-53]. Alginate is generally extracted from species like Macrocystis pyrifera, Ascophyllum nodosum and Laminaria hyperborea [51,52]. Also, a few strains of genera Azotobacter (Azotobacter vinelandii) and mucoid strains of Pseudomonas (Pseudomonas aeruginosa) produce alginate [54,55]. Alginates are made up of different amounts of the C-5 epimer-L-guluronate (G) and D-mannuronate (M), which are joined by 1-4 glycosidic linkages. The source of the alginate affects the M/G residue ratio and, consequently, the material’s characteristics [54]. Alginate’s chemical composition differs between genera. The alginate’s molecular weight typically ranges from 500 kDa to 1,000 kDa [55]. For cells, immobilization in an insoluble alginate matrix is acknowledged as a quick and safe technique [56-57]. Torres et al. immobilized Pseudomonas fluorescens in Ca-alginate to successfully remove various kinds of phenols (phenol, chlorophenol, dichlorophenol and trichlorophenol) from wastewater. Polyak et al. immobilized the bioluminescent reporter cells which were enclosed in microspheres using the novel biotin- alginate conjugate [53]. These biotinylated alginate microspheres were used to create a biosensor, which was then used to determine the concentration of the antibiotic mitomycin C, a model toxin, on the surface of an optical fiber coated with streptavidin.
Agar and Agarose
Agar which is also known as agar-agar is a strong gelling phycocolloid. It is produced by various genera of marine red algae like Pterocladia, Gigarti no, Gelidium and Gracilaria. Its primary chemical composition consists mostly of repeating units of D-galactose and 3-6, anhydro- L-galactose with little variation and a low concentration of sulphate esters. It is a polysaccharide which is formed in agarophyte algae’s cell walls. Depending on the raw material and the production procedure used, agar is a combination of agarose and agaropectin fractions in varying quantities. Solely, the agarose component in agar causes it to gel. It melts at 96° and turns into jelly when it cools to 40° or 45°. Cells can be trapped in agar in the form of sphere-shaped beads, blocks, or membranes [58-61]. Kim et al. developed a biosensor that could identify the toxicity of hydrogen peroxide, phenol, and mitomycin C in water samples when four bioluminescent Escherichia coli strains were immobilized in a 96- well plate with an LB-agar matrix.
The components of agar that gels are called agarose. They have low sulphate contents, often below 0.15%, and high molecular weight of>100,000 Daltons and commonly exceeding 150,000 Daltons. Once agar deposits in the cell wall of red algae, it undergoes enzymatic polymerization and desulfonation, converting majorly to agarose. Agarose creates “physical gels,” in which the structure of these aqueous gels is entirely made up of polymer molecules, which combine through hydrogen bonds. Reversibility of the ‘physical gels’ is the most notable characteristic. On heating, it is in the dissolved liquid form, while on cooling it forms gel. Gracilariopsis chorda (i.e. agar agar), is often treated with alkali to produce agarose [59,62,63].
As an example of its application, for the study of Biochemical Oxygen Demand (BOD) in wastewater from the pulp and paper industries with a high cellulose concentration, Raud et al. developed two semi-specific bacterial biosensors [64]. In this biosensor, cellulose degrading bacterial cells (Bacillus subtilis and Paenibacillus sp.) were immobilized in an agarose gel matrix which served as the backbone for the biosensor. Fifteen days following immobilization, the steady phase permitting reproducible measurements of BOD began. Biosensors had a 96-day service life before an inconsistent sensor signal was noticed. The stability of the immobilized microorganisms has a significant impact on the operational stability as well as on the service life of BOD biosensors [64]. This indicates a very good example of biosensor with whole-cell immobilized system.
Polyvinyl alcohol
PVA is a biodegradable, water-soluble vinyl polymer made exclusively of carbon-carbon bonds [65,66]. In several biological systems, PVA has been thoroughly investigated as a matrix for cell immobilization [67]. It is the only industrially manufactured vinyl polymer that is known to be mineralized by microorganisms. A bacterial strain called Pseudomonas O3 degrades PVA and uses it as source of both carbon and energy. Iterative freeze-thaw gelation of PVA is possible at a temperature of -20°, which is a very gentle way of gelation [68].
Using it Philp et al. immobilized two bioluminescent strains. One bacterial strain was natural (Photobacterium (Vibrio) fischeri) and the other was genetically modified (Pseudomonas putida BS566: luxCDABE). Both the bacterial strains were compared after immobilization on thin films of polyvinyl alcohol. The biosensors were evaluated in comparison to pure phenol and 3-chlorophenol, a reference hazardous chemical that is well-known to be far more harmful to bacteria than phenol. The industrial effluent containing phenolics was then used to test the toxicity through biosensors.
Polyacrylamide
Polyacrylamides (PAMs) are synthetic linear polymers consisting of acrylamide or a mixture of acrylamide and acrylic acid. PAMs are water-soluble and inexpensive polymers. Polyacrylamide was the first artificial gel to be used to entrap live microbial cells [31,69]. Chen et al. immobilized oil-degrading bacteria (Pseudomonas X1, Acinetobacter D1, Bacillus A1 and A2) in a mixture of polyacrylamide and sodium-alginate beads [70]. Liu et al. proposed that polyacrylamide (PAM-Alginate) hydrogel could provide sufficient water, oxygen and nutrients to the whole-cell biosensors [71]. Genetic circuits were constructed in E. coli DH5α to detect cognate inducers (i.e., DAPG (2, 4-diacetylphloroglucinol), AHL (N-acyl homoserine lactone), IPTG (isopropyl β-D-1-thiogalactopyranoside), and Rham (rhamnose)) and express gfp (Table 1).
| Immobilization matrix | Organism immobilized | Analyte | Type of detection | Reference |
|---|---|---|---|---|
| Glass particles | Pseudomonas putida | Phenolic compounds | - | [72] |
| pre-treated with polyethylene diamine | ||||
| Strontium-alginate | Pseudomonas fluorescens HK44 | Naphthalene and Salicylate | Optical | [25] |
| Agarose gel | Acinetobacter baylyi ADPWH_recA | Cadmium | Diffusive Ggradients in Thin-films (DGT) Technique | [73] |
| Agarose | E. coli DH1 | Cadmium | Optical | [74] |
| pBzntlux | As2O3 | |||
| pBarslux | - | |||
| pBcoplux and | - | |||
| E. coli XL1 pBfiluxCDABE | - | |||
| Polyurethane | Escherichia coli | Mercury | Optical | [75] |
| RBE27-13 | ||||
| Agarose | Escherichia colistrain DH5α | Phenolic compounds | ELISA plate reader (Versa Max, VA, USA) | |
| Agarose | Escherichia colistrain DH5α | Hg2+, Cu2+, and Cd2+ | - | [77] |
| Agarose | E. coli DH5α 2296 | Aromatic hydrocarbon compounds (Sodium benzoate, Naphthalene) | Spectro-fluorometer | [18] |
| Calcium alginate | E. coli DH5α 2301 | |||
| Gelatin | - | |||
| Alginate beads | - | |||
| Agar | E. coli MG1655(pBR-arsR773) | Arsenite | Optical based Multiskan GO UV/ | [78] |
| Alginate beads | Arsenate | Vis. Microplate | ||
| Agarose | Bacillus subtilis | Arsenic | Plate reader, Varioskan Lux 3020-428 (Thermo Scientific) | [79] |
| Optical based |
Note: DH5α- ; .
Table 1: Immobilization of cells on different matrices in biosensors.
Approaches towards integration of whole-cell biosensing strains in a detection device [72-79].
Advances in recent synthetic biology, material sciences and bio-electronics have paved the way for the design and fabrication of innovative whole-cell deployable biosensor devices [80]. Generally, there are three components of biosensors as shown in Figure 8. A: Primarily, a substrate that is the key element in the whole system; B: Bioreceptor-as biorecognition element specific to the required analyte; C: a signal from the bioreceptor given to the transducer which is then converted into readable form by the user [81].
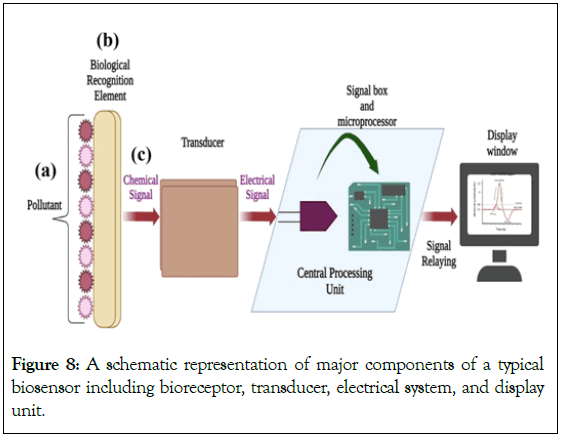
Figure 8: A schematic representation of major components of a typical biosensor including bioreceptor, transducer, electrical system, and display unit.
The design contains genetically engineered microbes as the core followed by an electronic detection system [82]. In whole-cell biosensors, two transducers are involved. The principal transducer, which is an integral part of the cell responding to environmental signals, is in the form of fluorescent protein synthesized. While the secondary transducer- an optical or electrochemical detection system, will measure and record the primary transducer’s intensity [83]. The current challenge is the fabrication of a whole-cell based biosensing device that satisfies all the requirements for directly interacting with biological systems. The challenge is not just limited to the sensing part but to the other crucial aspects such as fluid delivery, analyte area, zone of signal detection and mainly interconnecting the whole system on a single platform [84]. Another aspect that plays a role is the flexibility of the biosensing device and the viability of the organisms for direct application. Due to the limited understanding of the interaction between the device and bio-interface the attachment of those biosensing strains needs more attention [85].
Here, we review the latest developments of whole-cell biosensor devices followed by the challenges and perspectives towards their broad applications in environment. In the last few decades, significant research has been carried out in employing whole-cell biosensors where efforts were majorly focused towards its design and construction for the detection of a wide range of pollutants [82]. There is plenty of proof as stated in the review later, for: the successful integration of biosensing strains into microstructures; signal transduction by detectors; and preservation of biosensing strains in a way permitting continuous measurements. Several examples of single measurement and multiple parallel measurements have been described here on microfluidic-based sensing systems [86,87]. As a proof of concept, Buffi and his colleagues developed and tested a microfluidic chip which was composed of a Polydimethylsiloxane (PDMS) block containing the designed channels and cages in which biosensing strains E. coli having gfp as reporter gene was encapsulated in agarose beads and retained in the chip [88]. The chip had two parallel fluidic lines and cages for duplicate measurements of the sample. Biosensing strains were stored at -20° for a period of one month which retained inducibility. The system also estimated the number of biosensing strains in beads through the light scattering that permitted signal amplification that was detected in a range of 0-50 mg L-1 concentration of arsenic with an exposure time of 200 min. Despite the success, the essential difficulty faced in the single- use of disposable chips was signal calibration.
Concerning multiple parallel detections for different targets, work of Tsai and his colleagues was tremendously appreciated [89]. A 16-well aluminium microfluidic chip (Lumichip) with an oxygen- permeable flow channel was developed which contained different biosensing strains sensing different toxicants. The Lumichip was comprised of 3 parts: a 2 mm thick PDMS (Polydimethylsiloxane) lid; a 2.5 mm aluminium chip; and an optically clear film for sealing the bottom of the aluminium chip. The optical system setup consisted of 2 subsystems: the temperature control subsystem [31], and the CCD [32]. The temperature control subsystem contained a Proportional Integral Derivative (PID), a thermocouple attached to an aluminium chip, and a transformer for resistive heating. The Lumichip was then placed on a Charge-Coupled Device (CCD) sensor (lens-free imaging configuration) to detect the time-lapse changes emitted by the biosensing strains. The integration of each good position was then acquired by the Lumilogger program followed by the automated custom program Python.
Elad et al. developed a bacterial bioluminescent biosensor in which the bacteria was immobilized in agarose for the continuous monitoring of water toxicity posed by Chemical industry (Table 2) [90].
| Type of transduction device | Organisms | Type of pollutant | Limit of detection |
|---|---|---|---|
| Electrochemical miniaturized silicon | E. coli DH5α | As (III) | 1.5 ppb |
| Hg (II) | 0.1 ppb | ||
| Optical SiPM (silicon photomultiplier) | E. coli DH5α | Hg (II) | 0.25 µg/L |
| Field-Ready Electrochemical Devices (FRED-As) | E. coli DH5α | As (III) | 2.2 ppb |
| Electrochemical by using eDAQ potentiostat (e-corder 410, Denistone East, Australia) | Bacillus sp. | Phenol | 3.0 ng/ L |
| Optical-electrochemical (Microplate based fluorescence) | E. coli-roGFP2 | Naphthalene | 1 × 10-4 ppm |
| Arsenite | 1.0 × 10-7 ppm | ||
| Cu2+ | 1.0 × 10-4 ppm | ||
| Cd2+ | 1.0 × 10-3 ppm | ||
| Optical fiber | Fluorescent E. coli RBE27-13 (pECFP) | Hg2+ | 100 nM (1 h) |
| Optoelectronic (disposable card) | E. coli::lux AB TBT3 | Tributyltin | 0.08 μM (1 h) |
| Optical (multi-well removable card) | E. coli DH1 pBzntlux, pBarslux, pBcoplux, and E. coli XL1 pBfiluxCDABE | CdCl2 | 0.5 μM |
| As2O3 | 5 μM | ||
| Ultra low-light CMOS bio-imager sensors (Complementary metal oxide semiconductor) | E. coli TV1061 | Formaldehyde | 1×10-8 mol L-1 |
| Optical fiber spectrophotometer | Flavobacterium sp. MTCC 2495 | Methyl parathion | 0.3 μM |
Table 2: Various types of transduction devices for the detection of pollutants with its Limit of Detection (LOD) with whole-cell biosensing strains.
The model chemicals compounds used in this study were nalidixic acid sodium salt, mitomycin C, methylviologendichlorid hydrate (paraquat), menadione sodium bisulfite, sodium (meta) arsenite, and potassium antimony (III) tartrate hydrate [91-99]. Based on the flow-through technique, the system was comprised of 4 flow-through chambers each made up of glass, a PDMS chip, and a polymethyl methacrylate cover. Parallel three chips containing 12 wells were loaded with immobilized biosensing strains and the sample was made to flow by multichannel peristaltic pump via detectors (three aligned single-photon avalanche diode device) for detection. Well-programmed LabVIEW (Laboratory Virtual Instrument Engineering Workbench) software was planned to control the detectors and quantify the generated signal. This whole system of flow through chambers-detectors was placed in a dark wooden box to ensure photon counting. These developed devices detected all simulated pollutants within 0.5 h to 2.5 h.
Philp and team [68], were focusing on monitoring industrial wastewater containing phenolics as contaminants. The system was designed by using genetically modified biosensing strains. The system contained two silicon photodiodes (450 mV µW−1 cm−2) as transducers and amplifiers for temperature drift. One photodiode was taken to detect the light signal emitted by biosensing strains, detecting contaminants, and another photodiode was taken to detect the light signal emitted by biosensing strains but without contaminants. Thus, the voltage output of the amplifier was observed to be proportional to the light signal difference due to the effect of contaminants only. The integrated system of PVA immobilization/photodiode transduction was abruptly giving results within 5 min.
A miniaturized whole-cell conductometric biosensor was constructed by Hnaien et al. for the detection of Trichloroethylene (TCE), a carcinogen from ground waters. The whole assembly was primed by using immobilized Pseudomonas putida F1 (PpF1) on 3D carbon nanotubes. The conductometric transducer of the sensor was comprised of two gold-layered microelectrodes on silicon dioxide and biosensing strains immobilized on it. Measurements of the sample were performed in Eppendorf tubes filled with 5 mM of phosphate buffer at optimum conditions of 23° temperature at pH 7.2. Both the microelectrodes were placed in the solution and kept until a steady signal output was obtained. Then they were removed and a sample was added to the tube followed by mixing and then the measurements were taken. The limit of detection of this biosensor for TCE was 0.07 µM (9 µg L-1) which is appropriate from an environmental perspective.
Apart from the microfluidic chip along with the electronic system, it is also now extensively accepted that optical-based biosensors with a combination of fluorescence and small molecules have attained greater success in terms of applications and sensitivity. Optic-based biosensors have become the foremost technology in biosensing involving fiber-optic chemistry [21]. Optical fibers are relatively low-priced and act as transduction mechanism. They are capable of exciting target molecules and capturing light from the emitted signal of the target molecule [99]. Regarding the portability of sensors, optic fiber-based biosensor relying on lensless contact imaging system (a Charge-Coupled Device (CCD)) was developed by Roda et al. and is presented in Figure 9. The CCD and the biosensing strain (E. coli JM109) immobilized on a multiwell polymeric matrix (to extend their shelf life) were combined to develop a compact portable device [100]. The entire system was constructed in a way so that trapped immobilized cells remains in direct contact with the CCD sensor via an optic fiber taper. The system was connected to a laptop for response and data. This developed system could detect lactose analogue isopropyl β-d-1- thiogalactopyranoside at nanomolar level with good accuracy and precision. The immobilized biosensing strains were stable up to one month. For multiplex detection, up to 4-8 bioluminescence signals could be obtained in a single analytical run.
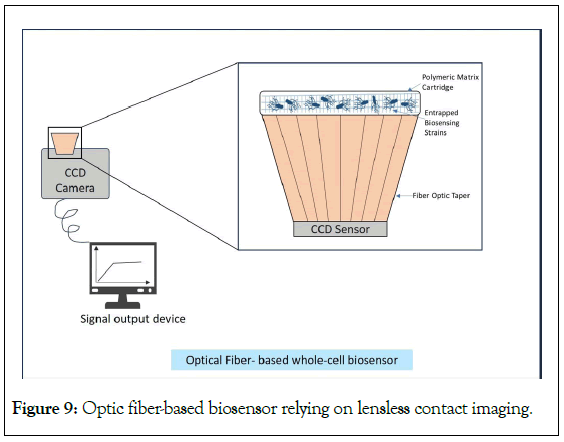
Figure 9: Optic fiber-based biosensor relying on lensless contact imaging.
Eltzov and his team developed an on-site photodetector device for detection of pollutants from wastewater [101]. The prototype was an integrated whole-cell biosensor consisting of biosensing strains (E. coli TV1061, E. coli DPD2544 and E. coli DPD2794) immobilized on a calcium alginate pad merged with optoelectronic system placed in a light-tight box as shown in Figure 10. The immobilized bacteria were placed above a liquid light guide (light transmission from cells to PMT) endface (53694, Edmund optic, USA) followed by a Photomultiplier Tube (PMT). A Hamamatsu HC135-01 PMT Sensor Module was used to measure bioluminescence (490 nm) with the sensitivity of PMT and the microcontroller. For data reviewing, software was developed using Lab View which allowed real-time monitoring of bioluminescent signals. For environmental testing, three different water samples were taken (Amazon, Sea of Galilee, and Ashdod rivers) and the biosensor was exposed to various pollutants such as organic solvents (toluene and formaldehyde), endocrine disruptive chemicals (atrazine, estron and bisphenol A) and heavy metals (zinc, arsenate, mercury and copper). It was summarized that the combination of immobilized biosensing strains and sensitivity of PMT module was highly convenient for the detection of these pollutants in wastewater.
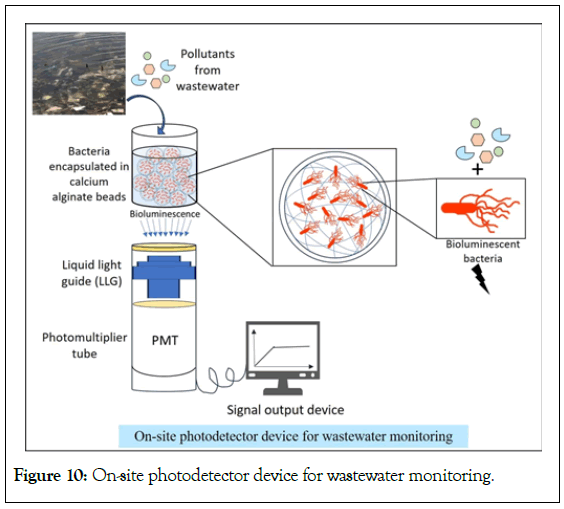
Figure 10: On-site photodetector device for wastewater monitoring.
Conclusion and Future Prospects
Biosensors are being used widely to detect the pollutants in the wastewaters. Considering whole-cell biosensors, from a product developing perspective, the aim is to safely hold cells within a sensor platform, letting them to relay a signal outside the immobilization matrix. In this review, an attempt is made to understand the system in three parts: construction of genetically modified biosensing strains; immobilization of cells on different matrices followed by release of signal; and an approach towards the device development. Designing of biosensing strain is carried out by fusion of a reporter gene and a promoter-operator of a pollutant degrading operon or ribosomal subunit. Such cloning is accomplished in a plasmid and then, it is transformed into the specific bacterial cells. These constructed cells can be immobilized by entrapping inside a polymer matrix, retained in the exceptional chambers of microfluidic devices, or merely by adsorption of cells to biocompatible substrates. Lastly for the measurements, PMT/liquid light guide like detectors can be used. The whole-cell biosensing systems have gained importance and attention due to its high efficiency which has resulted from the extensive field trials. Extensive studies are carried out in an attempt to reduce the number of handling steps for future field applications by combining advanced technologies in different ways to obtain precise results.
More focus has been placed in recent years on nanomaterial-based biosensors that demonstrate monitoring based on chemical reactions and biological phenomena that could be fascinating for the users. The small dimensions and large surface area of nanotechnologies, which improve surface activity and electrical conductivity, make them increasingly appealing and effective technologies. By merging well-established biological platforms with sophisticated engineering and cutting-edge product development technologies, integrated systems may be created that can meet the criteria for a user-friendly and convenient sensor development. Such systems when properly built can eliminate time-consuming sample preparation steps and challenges in the signal readout, making them convenient and reliable than traditional analytical procedures. The end product is expected to be a compact, mobile, field-ready system that can be made at an economically affordable cost and is simple enough to be used by someone with no prior experience.
Conflict of Interest
The authors declare no conflict of interest. Majority of the figures were created with Bio Render software.
References
- Asif S, Chaudhari A, Gireesh-Babu P, Chaudhuri PR, Sen R. Immobilization of fluorescent whole cell biosensors for the improved detection of heavy metal pollutants present in aquatic environment. Materials Today: Proceedings. 2016;3(10):3492-3497.
- Mukherjee A, Zaveri P, Patel R, Shah MT, Munshi NS. Optimization of microbial fuel cell process using a novel consortium for aromatic hydrocarbon bioremediation and bioelectricity generation. J Environ Manage. 2021;15(298):113546-113547.
[Crossref] [Google Scholar] [PubMed]
- Roy R, Ray S, Chowdhury A, Anand R. Tunable multiplexed whole-cell biosensors as environmental diagnostics for ppb-level detection of aromatic pollutants. ACS Sens. 2021;6(5):1933-1939.
[Crossref] [Google Scholar] [PubMed]
- Cao Y, Zhang B, Zhu Z, Xin X, Wu H, Chen B. Microfluidic based whole-cell biosensors for simultaneously on-site monitoring of multiple environmental contaminants. Front Bioeng Biotechnol. 2021;9 (9):622108. [Crossref]
[Google Scholar] [PubMed]
- Hashemi Goradel N, Mirzaei H, Sahebkar A, Poursadeghiyan M, Masoudifar A, et al. Biosensors for the detection of environmental and urban pollutions. J Cell Biochem.2018;119(1):207-212.
[Crossref] [Google Scholar] [PubMed]
- Woutersen M, Belkin S, Brouwer B, Van Wezel AP, Heringa MB. Are luminescent bacteria suitable for online detection and monitoring of toxic compounds in drinking water and its sources? Anal Bioanal Chem. 2011;915-929.
[Crossref] [Google Scholar] [PubMed]
- Su L, Jia W, Hou C, Lei Y. Microbial biosensors: a review. Biosens Bioelectron. 2011;26(5):1788-1799.
- Ekrami E, Pouresmaieli M, Shariati P, Mahmoudifard M. A review on designing biosensors for the detection of trace metals. Appl Geochemistry. 2021;1127:104902.
- Michelini E, Roda A. Staying alive: New perspectives on cell immobilization for biosensing purposes. Anal Bioanal Chem. 2012;402:1785-1797.
[Crossref] [Google Scholar] [PubMed]
- Mehta H, Patel P, Mukherjee A, Munshi NS. Biotechnological advances in detection of contaminants from wastewater. Clean (Weinh). 2023;51(3):2100439.
- Singh RL. Principles and applications of environmental biotechnology for a sustainable future. Springer Singapore. 2017.
- Ali J, Najeeb J, Ali MA, Aslam MF, Raza A. Biosensors: Their fundamentals, designs, types and most recent impactful applications: a review. J Biosens Bioelectron. 2017;8(1):1-9.
- Bose S, Maity S, Sarkar A. Review of microbial biosensor for the detection of mercury in water. Environ Qual Manag. 2022;31(4):29-40.
- Patel R, Zaveri P, Mukherjee A, Agarwal PK, More P, Munshi NS. Development of fluorescent protein-based biosensing strains: A new tool for the detection of aromatic hydrocarbon pollutants in the environment. Ecotoxicol Environ Saf. 2019;30(182):109450.
[Crossref] [Google Scholar] [PubMed]
- Bilal M, Iqbal HM. Microbial-derived biosensors for monitoring environmental contaminants: Recent advances and future outlook. Process Saf Environ Prot. 2019;124:8-17.
- Bjerketorp J, Håkansson S, Belkin S, Jansson JK. Advances in preservation methods: Keeping biosensor microorganisms alive and active. Curr Opin. Biotechnol. 2006;17(1):439.
[Crossref] [Google Scholar] [PubMed]
- Yoetz-Kopelman T, Dror Y, Shacham-Diamand Y, Freeman A. “Cells-on-Beads”: A novel immobilization approach for the construction of whole-cell amperometric biosensors. Sensors and Actuators B: Chemical. 2016;232:758-764.
- Patel R, Mukherjee A, Zaveri P, Mehta K, Aswani H, Munshi NS. Optimization of immobilization process and survival study of microbial sensing strains used for aromatic hydrocarbon detection in industrial wastewater. Water Environ J. 2020;34:937-948.
- Weibel DB, DiLuzio WR, Whitesides GM. Microfabrication meets microbiology. Nat Rev Microbiol. 2007;5(3):209-218.
[Crossref] [Google Scholar] [PubMed]
- Shacham-Diamand Y, Belkin S, Rishpon J, Elad T, Melamed S, Biran A, etal. Optical and electrical interfacing technologies for living cell bio-chips. Curr Pharm Biotechnol. 2010;11(4):376-383.
[Crossref] [Google Scholar] [PubMed]
- Vigneshvar S, Sudhakumari CC, Senthilkumaran B, Prakash H. Recent advances in biosensor technology for potential applications–an overview. Front Bioeng Biotechnol. 2016;4:11.
[Crossref] [Google Scholar] [PubMed]
- WN A, Chai MK, Wong LS. Whole-Cell Based Fluorescence Biosensors for Environmental Applications. Int J Mech Prod Eng. 2015;3(7).
- Divies C. Remarks on ethanol oxidation by an" Acetobacter xylinum" microbial electrode (author's transl). Ann Microbiol. 1975;126(2):175-186.
[Google Scholar] [PubMed]
- Sticher P, Jaspers MC, Stemmler K, Harms H, Zehnder AJ, Van Der Meer JR. Development and characterization of a whole-cell bioluminescent sensor for bioavailable middle-chain alkanes in contaminated groundwater samples. Appl Environ Microbiol. 1997;63(10):4053-4060.
[Crossref] [Google Scholar] [PubMed]
- Heitzer A, Malachowsky K, Thonnard JE, Bienkowski PR, White DC, Sayler GS. Optical biosensor for environmental on-line monitoring of naphthalene and salicylate bioavailability with an immobilized bioluminescent catabolic reporter bacterium. Appl Environ Microbiol. 1994;60(5):1487-1494.
[Crossref] [Google Scholar] [PubMed]
- Tecon R, Van der Meer JR. Bacterial biosensors for measuring availability of environmental pollutants. Sens. 2008;8(7):4062-4080.
[Crossref] [Google Scholar] [PubMed]
- Hu Q, Li L, Wang Y, Zhao W, Qi H, Zhuang G. Construction of WCB-11: A novel phiYFP arsenic-resistant whole-cell biosensor. J Environ Sci. 2010;22(9):1469-1474.
[Crossref] [Google Scholar] [PubMed]
- Guo M, Wang J, Du R, Liu Y, Chi J, He X, et al. A test strip platform based on a whole-cell microbial biosensor for simultaneous on-site detection of total inorganic mercury pollutants in cosmetics without the need for predigestion. Biosens Bioelectron. 2020;150:111899.
[Crossref] [Google Scholar] [PubMed]
- Kim HJ, Lim JW, Jeong H, Lee SJ, Lee DW, Kim T, et al. Development of a highly specific and sensitive cadmium and lead microbial biosensor using synthetic CadC-T7 genetic circuitry. Biosens Bioelectron. 2016;79:701-708.
[Crossref] [Google Scholar] [PubMed]
- Kumar J, Jha SK, D'souza SF. Optical microbial biosensor for detection of methyl parathion pesticide using Flavobacterium sp. whole cells adsorbed on glass fiber filters as disposable biocomponent. Biosens Bioelectron. 2006;21(11):2100-2105.
[Crossref] [Google Scholar] [PubMed]
- Willaert R. Cell immobilisation and its applications in biotechnology: Current trends and future prospects. In Fermentation microbiology and biotechnology, second edition. 2006:289-362.
- Zhu Y. Immobilized cell fermentation for production of chemicals and fuels. 2007:3733-3796.
- Dervakos GA, Webb C. On the merits of viable-cell immobilisation. Biotechnol Adv. 1991;9(4):559-612.
[Crossref] [Google Scholar] [PubMed]
- Bouabidi ZB, El-Naas MH, Zhang Z. Immobilization of microbial cells for the biotreatment of wastewater: A review. Environ Chem Lett. 2019;17:241-257.
[Crossref] [Google Scholar] [PubMed]
- Stolarzewicz I, Białecka-Florjańczyk E, Majewska E, Krzyczkowska J. Immobilization of yeast on polymeric supports. Chem BiochemEng. Q. 2011;25(1):135-144.
- Jack TR, Zajic JE. The immobilization of whole cells. Adv Biochem Eng. 2005;125-145.
- Burgain J, Gaiani C, Linder M, Scher J. Encapsulation of probiotic living cells: From laboratory scale to industrial applications. J Food Eng.2011;104(4):467-483.
- Trelles JA, Rivero CW. Whole cell entrapment techniques. Methods Mol Biol. 2013;365-374.
[Crossref] [Google Scholar] [PubMed]
- Martins SC, Martins CM, Fiúza LM, Santaella ST. Immobilization of microbial cells: A promising tool for treatment of toxic pollutants in industrial wastewater. Afr J Biotechnol. 2013;12(28).
- Górecka E, Jastrzębska M. Immobilization techniques and biopolymer carriers. Biotechnol Food Sci. 2011;75(1):65-86.
- Quan LM, Khanh DP, Hira D, Fujii T, Furukawa K. Reject water treatment by improvement of whole cell anammox entrapment using polyvinyl alcohol/alginate gel. Biodegradation. 2011;22:1155-1167.
[Crossref] [Google Scholar] [PubMed]
- Zommere Z, Nikolajeva V. Immobilization of bacterial association in alginate beads for bioremediation of oil-contaminated lands. Environ Exp Bot. 2017;15:105-111.
- Patel Y, Gupte A. Biological treatment of textile dyes by agar‐agar immobilized consortium in a packed bed reactor. Water Environ Res. 2015;87:242-251.
[Crossref] [Google Scholar] [PubMed]
- Jesionowski T, Zdarta J, Krajewska B. Enzyme immobilization by adsorption: A review. Adsorption. 2014;20:801-821.
- Bayat Z, Hassanshahian M, Cappello S. Immobilization of microbes for bioremediation of crude oil polluted environments: A mini review. Open Microbiol J. 2015;9:48.
[Crossref] [Google Scholar] [PubMed]
- Ramakrishna SV, Prakasham RS. Microbial fermentations with immobilized cells. Current science. 1999;10:87-100.
- Martinkova P, Kostelnik A, Valek T, Pohanka M. Main streams in the construction of biosensors and their applications. Int J Electrochem. 2017;12(8):7386-7403.
- Cassidy MB, Lee H, Trevors JT. Environmental applications of immobilized microbial cells: A review. J Ind Microbiol Biotechnol. 1996;16:79-101.
- Ma J, Harpaz D, Liu Y, Eltzov E. Smartphone-based whole-cell biosensor platform utilizing an immobilization approach on a filter membrane disk for the monitoring of water toxicants. Sensors 20. 20(19):5486.
[Crossref] [Google Scholar] [PubMed]
- Smidsrod O, Braek G. Alginate as immobilization matrix for cells. Trends Biotechnol. 1990;8:71-78.
[Crossref] [Google Scholar] [PubMed]
- Szekalska M, Puciłowska A, Szymańska E, Ciosek P, Winnicka K. Alginate: Current use and future perspectives in pharmaceutical and biomedical applications. Int J Polym Sci. 2016.
- Roy I, Gupta MN. Bioaffinity immobilization. Immobilization of Enzymes and Cells. 2006:107-116.
- Polyak B, Geresh S, Marks RS. Synthesis and characterization of a biotin-alginate conjugate and its application in a biosensor construction. Biomacromolecules. 2004;5(2):389-396.
[Crossref] [Google Scholar] [PubMed]
- Hay ID, Rehman ZU, Moradali MF, Wang Y, Rehm BH. Microbial alginate production, modification and its applications. Microb Biotechnol. 2013;6(6):637-650.
[Crossref] [Google Scholar] [PubMed]
- Sachdeva A, Gupta V, Rahi RK, Neelam D. Seaweed polysaccharides based edible coatings and films: An alternative approach. Education. 2015;2020.
- Melvik JE, Dornish M. Alginate as a carrier for cell immobilisation. Springer. 2004:33-51.
- Torres LG, Sánchez-De-La-Vega A, Beltran NA, Jiménez BE. Production and characterization of a Ca-alginate biocatalyst for removal of phenol and chlorophenols from wastewaters. Process Biochem. 1998;33(6):625-634.
- Rathnayake IV, Megharaj M, Naidu R. Green fluorescent protein based whole cell bacterial biosensor for the detection of bioavailable heavy metals in soil environment. Environ Technol Innov. 2021;23:101785.
- Armisen R, Gaiatas F. Agar. InHandbook of hydrocolloids 2009:82-107. Woodhead Publ.
- Mahajan R, Gupta VK, Sharma J. Comparison and suitability of gel matrix for entrapping higher content of enzymes for commercial applications. Indian J Pharm Sci. 2010;72(2):223.
[Crossref] [Google Scholar] [PubMed]
- Kim BC, Gu MB. A bioluminescent sensor for high throughput toxicity classification. Biosens. Bioelectron. 2003;18(8):1015-1021.
[Crossref] [Google Scholar] [PubMed]
- Jiang B, Song Y, Liu Z, Huang WE, Li G, Deng S, et al. Whole-cell bioreporters for evaluating petroleum hydrocarbon contamination. Crit Rev Environ Sci Technol. 2021;51(3):272-322.
- Nijenhuis K. Agarose. Thermoreversible Networks: Viscoelastic Properties and Structure of Gels. 1997;194-202.
- Raud M, Tutt M, Jõgi E, Kikas T. BOD biosensors for pulp and paper industry wastewater analysis. Environ Sci Pollut Res. 2012;19:3039-3045.
[Crossref] [Google Scholar] [PubMed]
- Doble M, Kumar A. Biotreatment of industrial effluents. Elsevier sci. 2005.
- Letcher TM, editor. Plastic waste and recycling: Environmental impact, societal issues, prevention, and solutions. Academic Press. 2020.
- Khoo KM, Ting YP. Polyvinyl alcohol as an immobilization matrix-a case of gold biosorption. Water Sci Technol. 2001;43(11):17-23.
[Crossref] [Google Scholar] [PubMed]
- Philp JC, Balmand S, Hajto E, Bailey MJ, Wiles S, Whiteley AS, et al. Whole cell immobilised biosensors for toxicity assessment of a wastewater treatment plant treating phenolics-containing waste. Anal Chim Acta. 2003;487(1):61-74.
- Moraskie M, Roshid MH, O'Connor G, Dikici E, Zingg JM, Deo S, et al. Microbial whole-cell biosensors: Current applications, challenges, and future perspectives. Biosens Bioelectron. 2021;191:113359.
[Crossref] [Google Scholar] [PubMed]
- Chen L, Zhao S, Yang Y, Li L, Wang D. Study on degradation of oily wastewater by immobilized microorganisms with biodegradable polyacrylamide and sodium alginate mixture. ACS omega. 2019;4(12):15149-15157.
[Crossref] [Google Scholar] [PubMed]
- Liu X, Tang TC, Tham E, Yuk H, Lin S, Lu TK, et al. Stretchable living materials and devices with hydrogel–elastomer hybrids hosting programmed cells. Proc Natl Acad Sci. 2017;114(9):2200-2205.
[Crossref] [Google Scholar] [PubMed]
- Nandakumar R, Mattiasson B. A microbial biosensor using Pseudomonas putida cells immobilised in an expanded bed reactor for the online monitoring of phenolic compounds. Anal Lett. 1999;2379-2393.
- Wu S, Li H, Zhang D, Zhang H. Simultaneous determination of heavy metal concentrations and toxicities by diffusive gradient in thin films containing Acinetobacter whole-cell bioreporters (Bio-DGT). Environ Pollut. 2023;320:121050.
[Crossref] [Google Scholar] [PubMed]
- Charrier T, Durand MJ, Affi M, Jouanneau S, Gezekel H, Thouand G. Bacterial bioluminescent biosensor characterisation for on-line monitoring of heavy metals pollutions in waste water treatment plant effluents. Biosensors. 2010;302:233-244.
- Biran I, Rissin DM, Ron EZ, Walt DR. Optical imaging fiber-based live bacterial cell array biosensor. Anal Biochem.2003;315(1):106-113.
[Crossref] [Google Scholar] [PubMed]
- Shin HJ. Agarose-gel-immobilized recombinant bacterial biosensors for simple and disposable on-site detection of phenolic compounds. Appl Microbiol Biotechnol. 2012;93:1895-1904.
[Crossref] [Google Scholar] [PubMed]
- Kim Y, Choi H, Shin WH, Oh JM, Koo SM, Kim Y, et al. Development of colorimetric whole-cell biosensor for detection of heavy metals in environment for public health. Int J Environ Res Public Health. 2021;18(23):12721.
[Crossref] [Google Scholar] [PubMed]
- Elcin E, Öktem HA. Immobilization of fluorescent bacterial bioreporter for arsenic detection. J Environ Health Sci Eng. 2020;18:137-148.
[Crossref] [Google Scholar] [PubMed]
- Valenzuela-García LI, Alarcón-Herrera MT, Ayala-García VM, Barraza-Salas M, Salas-Pacheco JM, Díaz-Valles JF, et al. Design of a whole-cell biosensor based on bacillus subtilis spores and the green fluorescent protein to monitor arsenic. Microbiol Spectr. 2023:432-523.
[Crossref] [Google Scholar] [PubMed]
- Ziegler C, Göpel W. Biosensor development. Curr Opin Chem Biol.1998;2(5):585-591.
[Crossref] [Google Scholar] [PubMed]
- Xu M, Obodo D, Yadavalli VK. The design, fabrication, and applications of flexible biosensing devices. Biosens Bioelectron. 2019;124:96-114.
[Crossref] [Google Scholar] [PubMed]
- Pham HL, Ling H, Chang MW. Design and fabrication of field-deployable microbial bio sensing devices. Curr Opin Biotechnol. 2022;76:102731.
[Crossref] [Google Scholar] [PubMed]
- Flemming JH, Baca HK, Werner-Washburne M, Brozik SM, López GP. A packed microcolumn approach to a cell-based biosensor. Sens Actuators B Chem. 2006;113(1):376-381.
- Liu S, Chen G, Mei A, Lu X, Zhao F, Li Z, et al. Advanced microflow manipulation strategy in paper-based microfluidics towards smart analytical chemistry: A concise review. 2023.
- North SH, Lock EH, Taitt CR, Walton SG. Critical aspects of biointerface design and their impact on biosensor development. Anal Bioanal Chem. 2010;397:925-933.
[Crossref] [Google Scholar] [PubMed]
- Roggo C, van der Meer JR. Miniaturized and integrated whole cell living bacterial sensors in field applicable autonomous devices. Curr Opin Biotechnol. 2017;45:24-33.
- Song Y, Lin B, Tian T, Xu X, Wang W, Ruan Q, et al. Recent progress in microfluidics-based biosensing. Anal Chem.2018;91(1):388-404.
[Crossref] [Google Scholar] [PubMed]
- Buffi N, Merulla D, Beutier J, Barbaud F, Beggah S, Van Lintel H, et al. Development of a microfluidics biosensor for agarose-bead immobilized Escherichia coli bioreporter cells for arsenite detection in aqueous samples. Lab Chip. 2011;11(14):2369-2377.
[Crossref] [Google Scholar] [PubMed]
- Tsai HF, Tsai YC, Yagur-Kroll S, Palevsky N, Belkin S, Cheng JY. Water pollutant monitoring by a whole cell array through lens-free detection on CCD. Lab Chip. 2015;15(6):1472-1480.
[Crossref] [Google Scholar] [PubMed]
- Elad T, Almog R, Yagur-Kroll S, Levkov K, Melamed S, Shacham-Diamand Y, et al. Online monitoring of water toxicity by use of bioluminescent reporter bacterial biochips. Environ Sci Technol. 2011;45(19):8536-8544.
[Crossref] [Google Scholar] [PubMed]
- Sciuto EL, Petralia S, Van der Meer JR, Conoci S. Miniaturized electrochemical biosensor based on whole‐cell for heavy metal ions detection in water. Biotechnol Bioeng. 2021;118(4):1456-1465.
[Crossref] [Google Scholar] [PubMed]
- Sciuto EL, Coniglio MA, Corso D, Van der Meer JR, Acerbi F, Gola A, et al. Biosensors in monitoring water quality and safety: An example of a miniaturizable whole-cell based sensor for Hg2+ optical detection in water. Water. 2019;11(10):1986.
- Sánchez S, McDonald M, Silver DM, De Bonnault S, Chen C, LeBlanc K, et al. The integration of whole-cell biosensors for the field-ready electrochemical detection of arsenic. J Electrochem Soc 2021;168(6):067508.
- Ariyanti D, Iswantini D, Sugita P, Nurhidayat N, Effendi H, Ghozali AA, et al. Highly sensitive phenol biosensor utilizing selected Bacillus biofilm through an electrochemical method. J Sci.2020;24(1):4.
- Arias-Barreiro CR, Okazaki K, Koutsaftis A, Inayat-Hussain SH, Tani A, Katsuhara M, et al. A bacterial biosensor for oxidative stress using the constitutively expressed redox-sensitive protein roGFP2. Sensors. 2010;10(7):6290-6306.
[Crossref] [Google Scholar] [PubMed]
- Horry H, Charrier T, Durand MJ, Vrignaud B, Picart P, Daniel P, et al. Technological conception of an optical biosensor with a disposable card for use with bioluminescent bacteria. Sens Actuators B Chem. 2007;122(2):527-34.
- Axelrod T, Eltzov E, Marks RS. Bioluminescent bioreporter pad biosensor for monitoring water toxicity. Talanta. 2016;149:290-297.
[Crossref] [Google Scholar] [PubMed]
- Hnaien M, Lagarde F, Bausells J, Errachid A, Jaffrezic-Renault N. A new bacterial biosensor for trichloroethylene detection based on a three-dimensional carbon nanotubes bioarchitecture. Anal Bioanal Chem.2011;400:1083-1092.
[Crossref] [Google Scholar] [PubMed]
- Sohrabi H, Hemmati A, Majidi MR, Eyvazi S, Jahanban-Esfahlan A, Baradaran B, et al. Recent advances on portable sensing and biosensing assays applied for detection of main chemical and biological pollutant agents in water samples: A critical review. TrAC, Trends Anal Chem. 2021;143:116344.
- Roda A, Cevenini L, Michelini E, Branchini BR. A portable bioluminescence engineered cell-based biosensor for on-site applications. Biosens Bioelectron. 2011;26(8):3647-3653.
[Crossref] [Google Scholar] [PubMed]
- Eltzov E, Yehuda A, Marks RS. Creation of a new portable biosensor for water toxicity determination. Sens Actuators B Chem. 2015;221:1044-1054.
Citation: Munshi NS, Mehta H, Sachdeva A, Bhavsar J (2023) Exploring the Potential of Genetically Engineered Whole-Cell Biosensors: A Step towards Device Development for Environmental Monitoring. J Microb Biochem Technol. 15:558.
Copyright: © 2023 Munshi NS, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

