Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- ResearchBible
- Cosmos IF
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2020) Volume 9, Issue 5
Evaluation of Genetic Diversity in Aerial Yam (Dioscorea bulbifera L) using Simple Sequence Repeats (SSR) Markers
Aniefiok Ndubuisi Osuagwu* and Edem ULReceived: 19-Oct-2020 Published: 28-Dec-2020
Abstract
Dioscorea bulbifera is an underutilised crop of the family Dioscoreacea widely distributed throughout the tropical and sub Saharan regions of West Africa. Twenty five D. bulbifera accessions from West Africa held in International Institute of Tropical Agriculture (IITA) germ plasm bank were screened for genetic diversity using ten microsatellite loci in 25 ul volume reaction in a 96 well micro titre plate PCR reactions. The reaction mixture consisted of 3 µl of 10 ng/µl template DNA, 2µl of 2.5 mM DNTPs, 1 µl of 2.5 mM MgCl2, 1µ each of forward and reverse primers, 1 µl of DMSO4, 0.1 µl of 5 µg/µl Taq DNA polymerase (Invitrogen) and 3 µl of 10 ng/µl DNA. The total reaction volume was made up to 25 volume using 13.4 µl nuclease free water. The PCR programme consisted of denaturation at 94°C for 30 secs, followed by 35 cycles of 94°C for 30s, 55 or 45°C for 20s and 72°C for 30 s, with a final extension step at 72°C for 7mins. A total of 74 alleles were detected with an average allele number of 7.4 per locus. A Polymorphic Information Content (PIC) mean value of 0.74 showed existence of variability among the accessions. A mean gene diversity value of 0.77 was also observed. SSR approach proved to be a valuable tool in the determination of genetic diversity and relationships among D. bulbifera accessions. These results are significant in the conservation and genetic improvement program of this crop.
Keywords
Aerial yam; Genetic diversity; Conservation; Microsatellites; Polymorphic information content
Introduction
Dioscorea bulbifera which is also known as aerial yam is an edible yam species which although capable of providing nutrition for the hungry masses in some parts of the world where it occurs in abundance, has been largely neglected and underutilized. In the Tropical forest region of West Africa the plant occurs in several morphological forms and attempts are being made to characterize the species based on both the morphological attributes and in this report, using molecular tool such as Simple Sequence Repeats (SSR) marker. The aerial yam grows aggressively on diverse soil types often reaching up to 20m or more in length and yielding very many bulbils in one growth season. Miller, (2003) describes the plant as having stems that are round or slightly angled in cross section, with axillary, simple, fasciculate inflorescences consisting of diminutive and sessile flowers usually white or greenish tinged in colour [1].
Several landraces of D. bulbifera are cultivated for human consumption but some also occur in the wild and are not edible in Nigeria. Reports from ISSG (2012) indicate that some genotypes of Dioscorea bulbifera are rich in diosgenin, which is a useful bioactive substance in the production of some steroidal hormones, including some synthetic birth control pills, thus adding to the usefulness of this plant species and giving credibility to further study of its species complex for proper identification [2]. Adequate characterization of this plant especially at the molecular level will encourage and enhance its breeding for improvement of traits and utilization both for food and drug source. So far, lack of adequate knowledge of this food crop has significantly contributed to its genetic erosion.
DNA based markers have proven to be more reliable in the assessment of genetic diversity and relationships among crop species than just molecular studies. Beebe et al., reported that DNA markers are invaluable in cultivar identification due to the fact that neither plant phenology nor variable environmental conditions have any influence on them [3]. They are effective and efficient in differentiating among genotypes with identical morphological traits. Aggarwal et al., further noted that molecular markers are useful in phylogenetic relationship analysis among accessions as well as in true-to-type plant identification [4]. DNA markers are said to be effective and efficient if they are co-dominant, abundant and evenly distributed throughout the genome. They are highly polymorphic, highly reproducible, simple and quick to assay and enhance simple exchange of data between laboratories. Simple Sequence Repeats (SSR) markers fulfil most of these requirements, hence the choice of SSR markers in the present study. This study on the assessment of genetic diversity and relatedness of the aerial yam using SSR markers was however carried out on 25 accessions of D. bulbifera in IITA holding. This will provide an insight into the genetic diversity of the aerial yam species of the West African region since the crops in the IITA gene bank are collections from several Countries (Table 1).
| S/N | Accessions | Origin | Ecological zone |
|---|---|---|---|
| 1 | TDb-2857 | Equatorial Guinea | Forest |
| 2 | TDb-3045 | Nigeria | Forest |
| 3 | TDb-3049 | Benin | Savanna |
| 4 | TDb-3058 | Togo | Savanna |
| 5 | TDb-3060 | Togo | Savanna |
| 6 | TDb-3064 | Togo | Savanna |
| 7 | TDb-3067 | Togo | Savanna |
| 8 | TDb-3068 | Togo | Savanna |
| 9 | TDb-3069 | Togo | Savanna |
| 10 | TDb-3070 | Togo | Savanna |
| 11 | TDb-3072 | Nigeria | Forest |
| 12 | TDb-3078 | Nigeria | Forest |
| 13 | TDb-3082 | Nigeria | Forest |
| 14 | TDb-3083 | Gabon | Savanna wood land |
| 15 | TDb-3084 | Gabon | Forest |
| 16 | TDb-3085 | Nigeria | Forest |
| 17 | TDb-3089 | Equatorial Guinea | Forest |
| 18 | TDb-3431 | Nigeria | Forest |
| 19 | TDb-3512 | Togo | Forest |
| 20 | TDb-3693 | Congo | Forest |
| 21 | TDb-3694 | Congo | Forest |
| 22 | TDb-3835 | Nigeria | Swampy with tall tree and grasses |
| 23 | TDb-4119 | Guinea | Savanna |
| 24 | TDb-4120 | Sierra Leone | Savanna |
| 25 | TDb-4122 | Sierra Leone | Savanna |
Table 1: Accessions of D. bulbifera used in the study, their origin and ecological zone.
Literature Review
Dioscorea bulbifera is among the yam species in the genus Dioscorea, with great potentials in providing food and drugs for hungry masses of tropical Africa where it occurs in significant abundance. Limited efforts have however been made in genetic diversity studies in the genus Dioscorea, but knowledge of the biology of this crop is still incomplete
Detailed analysis of genotypes based on DNA analyses plays a critical role on species identification, genetic improvements and conservation of crops Some studies on yam have been carried out using various DNA marker systems; these include; RAPDs, AFLPs and SSRs [5-11]. Simple Sequence Repeats (SSR) markers are often preferred to other DNA marker systems in genetic diversity analysis because they are co-dominant and highly polymorphic and are thus, more informative [12,13].
In a study by Ramser et al. [14], 23 accessions of D. bulbifera collected across various ecological zones of Asia, Africa and Oceania were characterized using RAPD markers. Results from the study corroborated a previous report of the “independent domestication” of D. bulbifera in Asia and Africa. Tostain et al., carried out a genetic diversity evaluation among 146 accessions of D. rotundata collected from Benin using 10 microsatellite loci as well as the diversity analysis among 56 other accessions using 6 microsatellite loci [15]. There was an observation of a significantly high degree of heterozygosity. In yet another study, a distinct varietal group was observed among accessions originating from Cameroun which had a distinct clustering pattern from all other West African accessions [16]. The clustering pattern suggested the uniqueness of the gene pool of this group that may be exploited for breeding and genetic improvement of Dioscorea genotypes in West Africa. Jayeola and Oyebola also reported the characterization of 34 accessions of D. bulbifera from Nigeria using morphological traits and SSR markers; with leaf colour and shape, petiole length, bulbil size and shape significantly contributing to variations among the populations studied [17].
Materials and Methods
This is the report of a research carried out in the germplasm bank and the Bioscience Centre of the International Institute of Tropical Agriculture (IITA), Ibadan-Oyo State, Nigeria.
Twenty five accessions of Dioscorea bulbifera collected from eight countries and maintained in IITA germplasm bank were used for the study (Table 1). All accessions were planted in 30cm size pots filled with sterilized top soil and maintained in a screen house at IITA, Ibadan, Nigeria.
DNA isolation and quantification
Cetyl Trimethyl Ammonium Bromide (CTAB) method was used to isolate Genomic DNA from fresh young leaves. The quality and concentration of DNA was assessed by gel electrophoresis using 8% agarose in1XTBE, cooled to 6°C and and 5 ul safe view pouredon gel tray. Gel was run at 80 Volts for 6 mins. Nano-drop spectrophotometer (Beckman Coulter DU 530) was used for quantification of DNA at 260 nm. Purified DNA for all samples was diluted in water to obtain DNA concentration of 25 ng/µL.
Polymerase chain reaction (PCR) and fragment analysis
A total of ten SSR primer pairs were used in the study (Table 2). An automated thermal cycler (Peltier thermal Cycler 200) was used to conduct the PCR reactions in a 25 µl volume in a 96- well microtitre plate. The reaction mixture consisted of 2.5 ul of 10x PCR buffer, 3ul of 10 ng/µl template DNA, 2 µl of 2.5 mM dNTPs, 1 µl of 2.5 mM MgCL2, 1 µl each of forward and reverse primers,1µl of DMSO4 1x reaction buffer and 0.1 µl of 5 u/µl Taq DNA polymerase (Invitrogen). The total reaction volume was made up to 25 µl using 13.4 µl nuclease free water.
| Name | Forward and reverse sequences | Repeat motif | Ta (°C) |
|---|---|---|---|
| Db2 | F: CACGACCTCCTGGAAGACAACT R: ATATAGCACGGGAGGCACAAAC |
(GAC)4 | 53 |
| Db3 | F: TTTTACCCAGGATTTAGAAGAA R: GGACTGGAGCCACAAGATT |
(CA)8 | 50 |
| Db5 | F: TGTCTATTATATTGCTCTTTCT R: CGTTTCTAATTTCTGGGTAT |
(GT)4 | 50 |
| Db6 | F: AAGCCGGTATCATTCAACAAAA R: CCCTCGCCAACATCAAGTAA |
(AC)8 | 53 |
| Db7 | F: CCGCAAGGCTCAAAAAGTTAGG R: TCGTGGATGAAGATGGGTGGAC |
(GA)4 | 53 |
| Db8 | F: TCCCAAGAAATCCAGAATA R: ATGCATGCCAAAACAAATA |
(GAC)4 | 50 |
| DBSSR1 | F: ACACACACACACAGAGAGAGAG R: AGAAGTTTGTTGCCCGTC |
(AC)6 (AG)20… (GGA)3 | 54 |
| DBSSR2 | F: ACACACACACACAGAGAGAGAG R: AACGCATCCCACCACTTC |
(AC)6 (AG)13 | 54 |
| DBSSR3 | F: ACACACACACACAGAGAGAGAG R: CACGATGGAGGAACACTT |
(AC)6 (AG)19 | 54 |
| DBSSR4 | F: ACACACACACACAGAGAGAGAG R: GAAAAGGAGAAGCCGAAT |
(AC)6 (AG)9 | 54 |
| DBSSR5 | F: ACACACACACACAGAGAGAGAG R: TTGATTGAAAAGGGAGGCT |
(AC)6 (AG)9 | 54 |
Ta =Annealing temperature
Table 2: Characteristics of ten microsatellite primers used to characterize 25 accessions of D. bulbifera.
The PCR cycling parameter consisted of a touch down PCR profile as follows; initial denaturation at 94°C for 5 mins, followed by 9cycles of denaturation at 94°C for 30 secs, annealing at 65°C for 30 secs, and elongation at 72°C for 30 secs followed by 35 cycles of denaturation at 94°C for 30 secs, annealing at 55C for 30 secs and elongation at 72°C for 30 secs. This was followed by a final extension step at 72°C for 7 min. and a holding temperature at 10°C. Amplified fragments were visualized on safe view stained 1.5% agarose gel electrophoresis. Visualization was further resolved on 6% polyacrylamide gel electrophoresis.
Data analysis
SSR fragment analysis: The base pairs (sizes) of fragments per accession were converted to binary data where alleles were transformed into presence (1) or absence (0) of an SSR band across the SSR markers. The genetic diversity indices such as number of alleles for locus, gene diversity and Polymorphic Information Content (PIC) were estimated using Power Marker version 3.25 software. The cluster pattern of the genetic diversity was constructed using DARwin 5.0 software. Analysis of Molecular Variance (AMOVA) was carried out using GenAIEX version 6.5.
Results
Genetic diversity indices analysis
Table 3 gives the summary of genetic diversity indices generated by 10 SSR markers. The largest number of alleles detected (11 alleles) was by DBSSR 2. This was followed by DB5 (10 alleles) DB3 and DBSSR4, respectively. Polymorphic Information Content (PIC) values were calculated to assess the discriminatory power of each SSR primer. A mean PIC value of 0.74 was obtained across the 10 SSR markers which ranged from 0.37 to 0.85 in loci DB7 and DBSSR4, respectively. Markers having lower allele frequency but higher number of alleles produced greater PIC as obtained in DBSSR4 (9 alleles and the highest PIC of 0.85, followed by DB5 with 10 alleles and PIC of 0.84, respectively. It can further be established from the results that marker DB7 had the highest allele frequency of 0.60 while markers DB3, DB5 and DBSSR4 had the lowest frequency of the major allele (0.24). The mean major allele frequency was 0.33 (Table 3).
| S/N | Marker | Major allele frequency | Allele no | Gene diversity | PIC |
|---|---|---|---|---|---|
| 1 | DB2 | 0.3600 | 8.0000 | 0.7936 | 0.7697 |
| 2 | DB3 | 0.2400 | 9.0000 | 0.8512 | 0.8345 |
| 3 | DB5 | 0.2400 | 10.0000 | 0.8544 | 0.8383 |
| 4 | DB6 | 0.3200 | 6.0000 | 0.7520 | 0.7105 |
| 5 | DB7 | 0.6000 | 2.0000 | 0.4800 | 0.3648 |
| 6 | DB8 | 0.3600 | 6.0000 | 0.7488 | 0.7095 |
| 7 | DBSSR2 | 0.2800 | 11.0000 | 0.8352 | 0.8176 |
| 8 | DBSSR3 | 0.3200 | 6.0000 | 0.7680 | 0.7314 |
| 9 | DBSSR5 | 0.3600 | 7.0000 | 0.7808 | 0.7521 |
| 10 | DBSSR4 | 0.2400 | 9.0000 | 0.8608 | 0.8460 |
| 11 | Mean | 0.3320 | 7.4000 | 0.7725 | 0.7374 |
Table 3: Genetic diversity indices for the ten SSR markers used to analyse 25 accessions of D. bulbifera.
High gene diversity was recorded, ranging between 0.48 in DB7 and 0.86 in DBSSR4 with a mean of 0.77. In general, the primers used were polymorphic as indicated by their allele frequencies which were all below 0.95 (Table 3). Plates 2 & 3 show SSR marker (DBSSR2 & DB5) profiles generated for the D. bulbifera accessions used in the study. In general, the unique migration pattern of the DNA fragments (bands) in the gel clearly discriminated the accessions.
Cluster analysis, dissimilarity coefficients and Analysis of Molecular Variance (AMOVA)
The genetic dissimilarity matrix of 25 accessions of D. bulbifera is given in Table 4. The result of the cluster analysis is given in a dendrogram in Figure 1.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 0.153846 | |||||||||||||||||||||||
| 3 | 0.132075 | 0.245283 | ||||||||||||||||||||||
| 4 | 0.166667 | 0.208333 | 0.183673 | |||||||||||||||||||||
| 5 | 0.428571 | 0.428571 | 0.44186 | 0.315789 | ||||||||||||||||||||
| 6 | 0.409091 | 0.454545 | 0.377778 | 0.35 | 0.352941 | |||||||||||||||||||
| 7 | 0.333333 | 0.25 | 0.346939 | 0.363636 | 0.578947 | 0.25 | ||||||||||||||||||
| 8 | 0.414634 | 0.414634 | 0.428571 | 0.405405 | 0.548387 | 0.515152 | 0.405405 | |||||||||||||||||
| 9 | 0.2 | 0.2 | 0.294118 | 0.173913 | 0.4 | 0.380952 | 0.26087 | 0.282051 | ||||||||||||||||
| 10 | 0.276596 | 0.276596 | 0.375 | 0.302326 | 0.513514 | 0.538462 | 0.348837 | 0.388889 | 0.2 | |||||||||||||||
| 11 | 0.395349 | 0.395349 | 0.454545 | 0.384615 | 0.454545 | 0.542857 | 0.384615 | 0.5625 | 0.317073 | 0.210526 | ||||||||||||||
| 12 | 0.5 | 0.458333 | 0.55102 | 0.5 | 0.473684 | 0.55 | 0.545455 | 0.621622 | 0.391304 | 0.348837 | 0.333333 | |||||||||||||
| 13 | 0.488372 | 0.44186 | 0.5 | 0.435897 | 0.515152 | 0.6 | 0.538462 | 0.625 | 0.414634 | 0.368421 | 0.235294 | 0.384615 | ||||||||||||
| 14 | 0.5 | 0.45 | 0.560976 | 0.444444 | 0.4 | 0.4375 | 0.5 | 0.655172 | 0.421053 | 0.428571 | 0.290323 | 0.333333 | 0.290323 | |||||||||||
| 15 | 0.319149 | 0.319149 | 0.416667 | 0.395349 | 0.513514 | 0.435897 | 0.348837 | 0.5 | 0.333333 | 0.333333 | 0.421053 | 0.348837 | 0.578947 | 0.4285714 | ||||||||||
| 16 | 0.55 | 0.55 | 0.560976 | 0.5 | 0.6 | 0.625 | 0.5 | 0.310345 | 0.368421 | 0.371429 | 0.354839 | 0.5 | 0.612903 | 0.5714286 | 0.428571 | |||||||||
| 17 | 0.423077 | 0.307692 | 0.471698 | 0.375 | 0.47619 | 0.5 | 0.416667 | 0.560976 | 0.32 | 0.361702 | 0.302326 | 0.333333 | 0.255814 | 0.4 | 0.404255 | 0.45 | ||||||||
| 18 | 0.44186 | 0.348837 | 0.545455 | 0.384615 | 0.454545 | 0.542857 | 0.435897 | 0.5625 | 0.365854 | 0.368421 | 0.294118 | 0.333333 | 0.235294 | 0.3548387 | 0.368421 | 0.54839 | 0.25581 | |||||||
| 19 | 0.391304 | 0.347826 | 0.404255 | 0.333333 | 0.444444 | 0.368421 | 0.285714 | 0.542857 | 0.363636 | 0.365854 | 0.243243 | 0.380952 | 0.297297 | 0.3529412 | 0.317073 | 0.47059 | 0.21739 | 0.18919 | ||||||
| 20 | 0.294118 | 0.333333 | 0.307692 | 0.361702 | 0.463415 | 0.395349 | 0.319149 | 0.5 | 0.306122 | 0.347826 | 0.333333 | 0.404255 | 0.380952 | 0.3846154 | 0.26087 | 0.53846 | 0.29412 | 0.33333 | 0.24444 | |||||
| 21 | 0.372549 | 0.333333 | 0.346154 | 0.319149 | 0.512195 | 0.44186 | 0.276596 | 0.5 | 0.306122 | 0.26087 | 0.190476 | 0.404255 | 0.333333 | 0.4358974 | 0.347826 | 0.38462 | 0.21569 | 0.28571 | 0.15556 | 0.16 | ||||
| 22 | 0.48 | 0.36 | 0.568627 | 0.434783 | 0.45 | 0.52381 | 0.478261 | 0.692308 | 0.416667 | 0.377778 | 0.365854 | 0.304348 | 0.317073 | 0.3684211 | 0.422222 | 0.63158 | 0.16 | 0.26829 | 0.36364 | 0.26531 | 0.26531 | |||
| 23 | 0.375 | 0.25 | 0.469388 | 0.363636 | 0.421053 | 0.45 | 0.409091 | 0.621622 | 0.347826 | 0.348837 | 0.333333 | 0.318182 | 0.333333 | 0.2777778 | 0.302326 | 0.61111 | 0.25 | 0.23077 | 0.28571 | 0.19149 | 0.23404 | 0.13043 | ||
| 24 | 0.589744 | 0.538462 | 0.55 | 0.485714 | 0.448276 | 0.548387 | 0.542857 | 0.428571 | 0.459459 | 0.411765 | 0.466667 | 0.428571 | 0.6 | 0.4074074 | 0.411765 | 0.33333 | 0.53846 | 0.46667 | 0.57576 | 0.47368 | 0.42105 | 0.45946 | 0.42857 | |
| 25 | 0.428571 | 0.387755 | 0.44 | 0.466667 | 0.487179 | 0.512195 | 0.511111 | 0.578947 | 0.404255 | 0.409091 | 0.4 | 0.466667 | 0.45 | 0.4594595 | 0.409091 | 0.62162 | 0.34694 | 0.45 | 0.44186 | 0.29167 | 0.33333 | 0.31915 | 0.28889 | 0.5 |
Table 4: Dissimilarity matrix among 25 accessions of D. bulbifera.
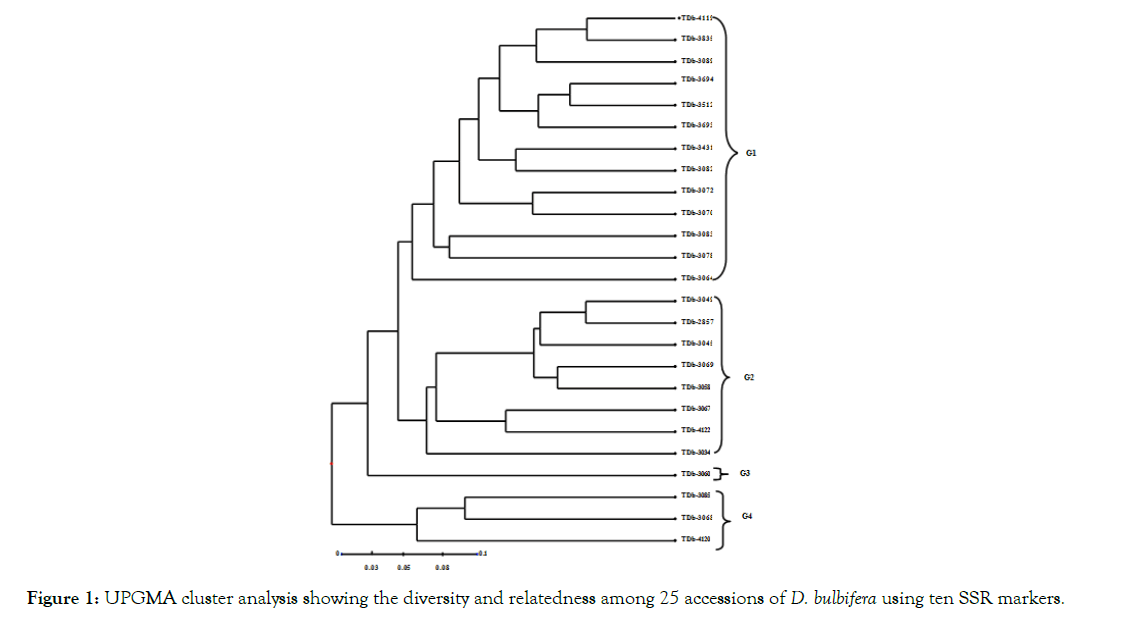
Figure 1: UPGMA cluster analysis showing the diversity and relatedness among 25 accessions of D. bulbifera using ten SSR markers.
The dendrogram revealed high intra-specific polymorphisms which enabled a reliable discrimination between accessions (Figure 1). The dendrogram revealed four main groups (G1, G2, G3 and G4) at a dissimilarity coefficient of 0.05. G1 contained 13 accessions including accessions from Guinea, Nigeria, Equatorial Guinea, Congo, Togo and Gabon. G2 contained eight accessions including 3 accessions from Togo and 1 each from Benin, Equatorial Guinea, Nigeria, Sierra Leone and Gabon. G3 contained only one accession (TDb-3060) from Togo. G4 contained 3 accessions, 1 accession each from Nigeria, Togo and Sierra Leone.
The least dissimilarity index was 0.13 (between TDb-3835 and TDb-4119) while the highest dissimilarity was 0.69 (between TDb- 3068 and TDb-3835) (Table 4). The mean dissimilarity index was 0.41 showing high intraspecific variation and polymorphism as evident in the clustering pattern (Figure 1). Analysis of Molecular Variance (AMOVA) for Simple Sequence Repeats of 25 accessions of D. bulbifera is summarized in Table 5. The result revealed 14% molecular variance among accessions collected from different geographical areas and 86% molecular variance among accessions within the areas of collection.
| SOV | DF | TSS | MSS | Estimated Variance | % mol. Variance | P -Value | Phqt |
|---|---|---|---|---|---|---|---|
| Among Pop. | 12 | 192.3 | 16.025 | 1.234 | 14 | 0.01 | 0.141 |
| Within Pop. | 35 | 203.89 | 5.82 | 8.775 | 86 | ||
| Total Pop. | 47 | 295.11 | 10.005 | 100 |
Key: phqt=Estimate of the population genetic differentiation based on permutation.
Table 5: Analysis of molecular variance (AMOVA) for simple sequence repeats of 25 accessions of D. bulbifera.
Discussions
The significant genetic variability observed in D. bulbifera germplasm from Equatorial Guinea, Nigeria, Benin, Togo, Gabon, Congo, Guinea and Sierra Leone could potentially be exploited for selection, breeding and genetic improvement schemes. The high genetic diversity or variation might be attributed to maintenance of heterologous chromosomal complex with high heterozygosity levels by D. bulbifera, in spite of being a vegetatively propagated crop. This was suggested by Siqueira et al., and may be confirmed through cytogenetic analysis of the yam chromosomes [18]. Obidiegwu et al., however suggested that the ancestry of some of the yam accessions could largely be determined by spontaneous hybridization since yams are dioecious, though the major contributing factor to variations exploited by farmers in crop improvement practices might be the selection of somatic mutants [19]. The high level of genetic diversity observed in D. bulbifera in this study might therefore be as a result of selection for somatic mutants arising from spontaneous hybridisations between dioecious aerial yam species.
Primer utility and efficiency were estimated by the number of alleles while the Polymorphic Information Content (PIC) indicated the differentiation capacity of each primer. Being a critical attribute of molecular markers, PIC could help to assess the discriminating capacity of the markers within the population [20]. Moghaddam et al., described PIC as a measure and assessment of the distribution of the frequencies of detected alleles [21]. Therefore, markers having lower allele frequency but higher allele number, had larger PIC as obtained in DBSSR4 (9 alleles with PIC of 0.85) as well as DB5 (10 alleles with PIC of 0.84), respectively. This indicated a better discrimination of the accessions. Norman et al., further describe PIC as an indicator of the ability to provide information which in turn is a function of the expected heterozygosity, usually calculated from allele frequencies. Such information is invaluable in selection, breeding and genetic improvement schemes [22]. All the 10 SSR loci were highly polymorphic in this study as indicated by high PIC values (0.74 on average). This implies that the SSR markers used in this study efficiently discriminated the D. bulbifera genotypes. The use of the PIC as an indicator of the discriminating power of a marker to differentiate among closely related individuals noted by Escandon et al., is thus affirmed [23].
A higher mean PIC value was observed in this study than what was reported in a study by Obidiegwu et al., on 89 accessions of D. alata where an average PIC value of 0.65 was recorded, using 13 SSR markers [19]. The result of this study might therefore indicate that D. bulbifera is a more diverse species than D. alata. This information could be invaluable in genetic diversity and phylogenetic relationship studies.
If the frequency of one of the alleles of a gene is between 0.95 and 0.99, such a gene is said to be polymorphic ([24]. Allelic frequency analysis results from this study showed high polymorphism across all the 10 primers used. Rare alleles i.e. alleles having frequencies below 0.l005, were not detected. This was possibly an indication of the adaptive nature of the D. bulbifera to the environmental/ soil types. Polymorphism enables and might be a result of gene reshuffling to enable adaptation to changing environments.
Cluster analysis based on the dissimilarity indices revealed thirteen groups (Figure 1). Each cluster had a fair representation of accessions from the different countries. The clustering pattern showed non distinction between country cultivars of D. bulbifera, this suggests wide distribution of clones. With the non-distinction between country cultivars, geographical location may therefore not have been a contributing factor to the distinction of the species. These results are consistent with the findings of Obidiegwu et al., in which SSR markers were used to characterize 89 accessions of D. alata from nine African countries and non-distinction between country cultivars of D. alata was reported. In a related study, AFLP analysis by Malapa et al., revealed the existence of three major groups of genotypes within D. alata, each assembling accessions from distant geographical origins and distinct ploidy levels.
Malapa et al., noted a wide distribution of the genotypes as clones over many years of human migration. According to the authors, there is a possibility of a common origin among some of the accessions. The authors further noted that most of the accessions which clustered together are most likely clones of a common source. Cluster analysis result from this study is thus consistent with the above observation, because most of the accessions from Nigeria and Togo which cluster together (Figure 1) may likely be clones with a shared ancestry.
The variation within clusters might be suggestive of high rate of mutations, many of which have effect on phenotype i.e. they are not neutral. Obidiegwu et al., opined that such mutations result in variations in phenotypic characteristics among the cultivars producing various shapes and colours for both aerial and underground organs. Analysis of Molecular Variance (AMOVA) for Simple Sequence Repeats of the 25 accessions of D. bulbifera (Table 5) revealed 14% molecular variance among geographical areas of collection of the accessions and 86% molecular variance within the areas of collection. It is evident from the result that 86% of the genetic variations in molecular data were attributable to variation at the DNA level in the different accessions while 14% of the variability was attributable to environmental influences and variation among the Dioscorea bulbifera populations. The result further corroborates the high genetic polymorphism indices obtained in this study.
A comparison of the clustering patterns in both morphological and SSR analysis for D. bulbifera in the present study showed little relationship. Such apparent lack of relationship between morphological and molecular data have previously been noted in several studies by different authors Montalvan et al., and supports the hypothesis that molecular polymorphisms are neutral in natural selection [25,26]. However, accessions TDb 3045 and TDb 3049 clustered together in the SSR analyses. Similarly, TDb 3512 and TDb 3693 were inseparable in the analyses. The SSR approach could be invaluable tool in the determination of germplasm diversity and genetic relationships among accessions and this is critical to the improvement of crops as well as in the taxonomy of species [27,28].
Conclusion
The 25 accessions of D. bulbifera assessed for genetic diversity using ten microsatellite loci showed 74 alleles with a mean of 7.4 alleles per locus. Polymorphic Information Content (PIC) mean value of 0.74 showed existence of variability among the accessions. There was non-distinction between country cultivars of D. bulbifera which suggested wide distribution of clones. Ultimately, SSR markers proved to be effective in characterizing the studied D. bulbifera germplasm. The study could help in improving the efficiency of D. bulbifera breeding programmes and cultivar development.
A combination of morphological characterization (previous report) and SSR approaches (in this report) proved to be viable tool in the determination of genetic diversity and relationships among 25 D. bulbifera accessions. This study is critical to the genetic improvement of the crop. Genetic variability was revealed in their clustering pattern on the dendrograms. Ultimately, this research has provided information on genetic diversity of D. bulbifera based on SSR markers and therefore serve as a baseline study and reference material for future research which will enable the development of proper conservation and breeding strategies for this species Distantly related accessions based on the SSR markers (TDb 3835 and TDb 3068, TDb 3060 and TDb 3085) should be exploited in breeding and genetic improvement scheme.
REFERENCES
- Miller JH. Nonnative invasive plants of southern forests: A field guide for identification and control. US Department of Agriculture, Forest Service, Southern Research Station. 2003.
- http://www.iucngisd.org/gisd/
- Beebe S, Skroch PW, Tohme J, Duque MC, Pedraza F, Nienhuis J. Structure of genetic diversity among common bean landraces of Middle American origin based on correspondence analysis of RAPD. Crop science. 2000;40(1):264-273.
- Agarwal M, Shrivastava N, Padh H. Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Rep. 2008;27(4):617-631.
- Asemota HN, Ramser J, Lopez-Peralta C, Weising K, Kahl G. Genetic variation and cultivar identification of Jamaican yam germplasm by random amplified polymorphic DNA analysis. Euphytica. 1995;92(3):341-351.
- Mignouna HD, Abang MM, Asiedu R. Harnessing modern biotechnology for tropical tuber crop improvement: Yam (Dioscorea spp.) molecular breeding. Afr J Biotechnol. 2003;2(12):478-485.
- Malapa R, Arnau G, Noyer JL, Lebot V. Genetic diversity of the greater yam (Dioscorea alata L.) and relatedness to D. nummularia Lam. and D. transversa Br. as revealed with AFLP markers. Genet Resour Crop Evol. 2005;52(7):919-929.
- Mignouna HD, Abang MM, Fagbemi SA. A comparative assessment of molecular marker assays (AFLP, RAPD and SSR) for white yam (Dioscorea rotundata) germplasm characterization. Ann Appl Biol. 2003;142(3):269-276.
- Scarcelli N, Tostain S, Vigouroux Y, Agbangla C, Daïnou O, Pham JL. Farmers’ use of wild relative and sexual reproduction in a vegetatively propagated crop: The case of yam in Benin. Mol Ecol 2006;15(9):2421-2431.
- Tostain S, Agbangla C, Scarcelli N, Mariac C, Dainou O, Berthaud J, et al. Genetic diversity analysis of yam cultivars (Dioscorea rotundata Poir.) in Benin using simple sequence repeat (SSR) markers. Plant Genet Resour. 2007;5(2):71-81.
- Silva DM, Siqueira MV, Carrasco NF, Montello CC, Nascimento WF, Veasey EA. Genetic diversity among air yam (Dioscorea bulbifera) varieties based on single sequence repeat markers. Genet Mol Res. 2016;15(2):615-728.
- Haussmann BIG, Geiger HH, Hess DE, Hash CT, Bramel-Cox P. Application of molecular markers in plant breeding. Training Manual Seminar, IITA, Ibadan-Nigeria. 2000.
- Gupta PK, Varshney RK. The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica. 2000;113(3):163-185.
- Ramser J, Weising K, Kahl G, López-Peralta C, Wetzel R. Genomic variation and relationships in aerial yam (Dioscorea bulbifera L.) detected by random amplified polymorphic DNA. Genome. 1996;39 (1):17-25.
- Tostain S, Scarcelli N, Brottier P, Marchand JL, Pham JL, Noyer JL. Development of DNA microsatellite markers in tropical yam (Dioscorea sp.). Mol Ecol. 2006;6(1):173-175.
- Mignouna HD, Ellis NT, Knox MR, Asiedu R, Ng QN. Analysis of genetic diversity in Guinea yams (Dioscorea spp.) using AFLP finger printing. Tropical Agriculture. 1998;75:24-28.
- Jayeola AA, Oyebola TO. Morpho-molecular studies in the natural populations of Dioscorea bulbifera in Nigeria. J Gen and Mol. 2013;14:1061-1128.
- Siqueira MM, Dequigiovanni G, Corazon-Guivin MA, Feltran JC, Veasey EA. DNA Finger printing of water yam (Dioscorea alata) cultivars in Brazil based in microsatellite markers. Hortic Bras. 2016;30:653-659.
- Obidiegwu JE, Asiedu R, Ene-Obong EE, Muoneke CO, Kolesnikova-Allen M. Genetic characterization of some water yam (Dioscorea alata L.) accessions in West Africa with simple sequence repeats. J Food Agric Environ. 2009;7(3&4):634-638.
- Ni J, Colowit PM, Mackill DJ. Evaluation of genetic diversity in rice subspecies using microsatellite markers. Crop Sci. 2002;42(2):601-607.
- Moghaddan M, Mohammadi SA, Mohebali’pour N, Toorchi M, Aharizad SA, Javidfar F. Assessment of genetic diversity in rape seed cultivars as revealed by RAPD and microsatellite markers. Afr J Biotechnol. 2009;8:3160-3167.
- Norman PE, Tongoona P, Shanahan PE. Diversity in chromosome number and raphide morphology of yam (Dioscorea spp.) genotypes from Sierra Leone. African Journal of Plant Science. 2011;6(4):157-160.
- Escandon AS, Zelener N, De La Torre MP, Soto S. Molecular identification of new varieties of Nierembergia linariaefolia (Graham) a native Argentinean ornamental plant. J Appl Genet. 2007;48(2):115-123.
- IPGRI & Cornell University. Genetic diversity analysis with molecular marker data. Learning Module. 2003;2-71.
- Montalvan R, Ando A, Echeverrigaray S. Use of seed protein polymorphism for discrimination of improvement level and geographic origin of upland rice cultivars. J Genet Mol Biol. 1998;21(4):1415-1590.
- Kimura M. Retrospective of the last quarter century of the neutral theory. Jap J Gen. 1993;68:521-528.
- Asiedu R, Ng SYC, Bai KV, Ekanayake IJ, Wanyera NMW. Genetic improvement Food yams: Advances in research. Ibadan: IITA and NRCRI. 1998;63-104.
- Martin FW. Tropical yams and their potential part 2. Dioscorea bulbifera. Agriculture Handbook, USDA. 1974;466.
Citation: Osuagwu AN, Edem UL (2020) Evaluation of genetic diversity in Aerial Yam (Dioscorea bulbifera L) Using Simple Sequence Repeats (SSR) Markers. Agrotechnology 9: 202. doi: 10.35248/2168-9881.20.9.202.
Copyright: © 2020 Osuagwu AN, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.


