Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- RefSeek
- Hamdard University
- EBSCO A-Z
- SWB online catalog
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2025) Volume 14, Issue 3
Estrogen Agonist Induced Effects on Platelet Activation Properties and Platelet Signaling Pathways by Structural Class
Veronica Mendes Soares1*, Amanda Mendes da Fonseca2, Otavio da Fonseca3, Moises Seabra3, Daniela Maria Alves Moreira Ramos3, Lorena Thayla Nascimento4 and Francisco das Chagas Alves Lima42Department of Education and Research, Hospital Alfa, Recife, Brazil
3Department of Geosciences, Federal University of Rio de Janeiro (UFRJ), Rio de Janeiro, Brazil
4Department of Chemistry, State University of Piauí, Teresina, PI, Brazil
Received: 08-Aug-2024, Manuscript No. PDS-24-26737; Editor assigned: 12-Aug-2024, Pre QC No. PDS-24-26737 (PQ); Reviewed: 26-Aug-2024, QC No. PDS-24-26737; Revised: 18-Jun-2025, Manuscript No. PDS-24-26737 (R); Published: 25-Jun-2025, DOI: 10.35248/2167-1052.25.14.391
Abstract
Background: The objective of this study is to identify the electronic structures of Platelet Receptors (RP) and the amino acid residues of the interaction of these receptors with estradiol and isoflavones through molecular anchoring involved in vascular thrombosis, directing investigations into their future clinical applicability.
Method: In an online database, G protein-coupled platelet receptors and tyrosine kinases involved in the coagulation process were identified. Bioinformatics tools (molecular anchoring) were used to characterize the interaction between each interaction site and bioactives (estradiol and isoflavones) in pre-clinical studies, evaluating the affinity between ligands and receptors.
Results: The molecular anchoring findings indicated that estradiol and isoflavones had good interaction with the receptors, revealing a risk of vascular thrombosis. Interestingly, estradiol showed interaction with both activating and inhibiting receptors in the platelet activation process and can be used as a pharmacological target.
Conclusion: In bioinformatics analysis, the platelet sites of the therapeutic mechanisms of estradiol and isoflavones were identified individually, as well as the intensity of each interaction that can be used in the production or modification of drugs to reduce the risk of vascular thrombosis in the treatment of menopausal symptoms.
Keywords
Platelet receptors; Thrombosis; Menopause treatment; Molecular anchoring
Highlights
- Understanding the chemical structure of the drug and its molecular mode of action, the relationship between the chemical structure and the molecular interactions between the receptor and its ligand leads to the development of new products.
- Bioinformatics analyzes biological data to more efficiently understand the implications and meaning of a variety of sequences and molecular structures and their interactions in clinical study.
Introduction
Innovation in the pharmaceutical industry with the development of new products starts from understanding the chemical structures of drugs and their molecular modes of action, the relationship between the chemical structure and the molecular interactions between receptors and their ligands. Translational research, understood as a process involved in the creation of knowledge, has been a topic of growing interest in the literature, and emerged with the objective of reducing the time between basic research and its clinical application, optimizing the time for results, reducing costs, integrating knowledge, promoting access to products for potential users, and enabling the practical application of knowledge generated in research with an impact on people's health, producing benefits for society [1-3]. Bioinformatics analyzes biological data to more efficiently understand the implications and meaning of a variety of sequences and molecular structures and their interactions [4].
Progressively, there is a growing need to convert knowledge from biological data through a computational approach. Recent bioinformatics tools are used to identify and characterize the active site of certain substances in preclinical studies, with an approach based on molecular anchoring revealing molecular targets quickly in discovering the role of platelets in various areas such as cell growth, infection, inflammation, inflammatory hemostasis, organ repair and thrombosis formation. Consequently, in-depth knowledge of platelet structure and function is becoming important in many fields of modern medicine, and a 'new' field of thrombo-inflammation is now an important driver of research into platelets with functions involving their receptors [5].
In thrombus formation, platelets are activated by various bloodborne vascular agonists, and a variety of platelet responses have been tested using measures of platelet aggregation from in vitro studies, where several platelet receptors could be identified.
During menopause, biological changes contribute to a greater risk of cardiovascular diseases. Several molecules used in hormone replacement to improve women's health conditions, such as estrogens associated or not with progestins, and phytochemicals such as isoflavones, with an effect on human physiology, have potentiated cardiovascular problems [6]. Therefore, in hormone replacement, due to the needs inherent to organic physiology and chronology, in which the risks and benefits of drug exposure must be analyzed, the identification of specific biological macromolecules and functional analysis must be previously evaluated.
Materials and Methods
In the study, bioinformatics tools and databases were used as central repositories to assist in the search for molecules and computational algorithms, following step-by-step instructions on how to use these tools.
Searches were carried out for active sites of the platelet G protein and tyrosine kinase enzymes for estrogen molecules and isoflavones, through virtual screening based on molecular anchoring between the selected enzymes and compounds. The necessary data were downloaded from the Protein Data Bank (PDB) structural database.
Obtaining the structures of I read
Using computational chemistry to describe the ground state, the bioactive molecular structures of the isoflavones daidzein, daidzin, genistein, genistin, glycitein, glycitin and the estrogen estradiol were optimized at the DFT (Density Functional Theory) level of theory using the computational package Gaussian. The B3LYP hybrid functional was combined with the 6-31+G (d,p) basis set. It is a hybrid functional with approximations most used in the application of description to large molecular systems, in which it was used to investigate the interaction of 17β-estradiol in the active site. The choice to use the method is due to its accuracy in predicting geometric parameters and the energy of large molecular systems with great precision, since such properties are crucial for studying a mechanism. reactional. Optimization is necessary to compare the theoretical geometric parameters of isoflavones, thus ensuring the quality of the physicochemical data obtained in computational calculations. The structures considered stationary points were subjected to frequency analysis and classified according to equilibrium geometry.
Anchoring molecular
Molecular interaction analysis between platelet receptors and the structures of EST and isoflavones-DAI, DAID, GEN, GENI, GLIC, GLI-with the enzymes glycoproteins G and platelet tyrosine kinase, computational simulations were carried out with the aid of the AutoDock program.
The AutoDock tool uses the scoring function of the interaction of the chemical compound and protein molecules according to the calculation of the binding energy (ΔG) based on the following formula:
ΔGbond=ΔGgauss+ΔGrepulsion+ΔGhbond+ΔGhydrophobic+ΔGtors.
ΔGgauss: Attractive term for the dispersion of two Gaussian functions; ΔGrepulsion: Square of the distance if closer than a threshold value; ΔGhbond: Ramp function-also used for interactions with metal ions; ΔGhydrophobic: Ramp function; ΔGtors: Proportional to the number of rotary connections [7,8] .
For the process, the estradiol and each isoflavone molecules were introduced into the active site of each platelet receptor enzyme and evaluated separately.
Each of the molecules was subjected to anchoring calculation in the following three steps: Step 1: Initially, the complexes of estradiol and each isoflavone were created, in each of the enzymes searched in the PDB (Protein Data Bank) database [9]; Step 2: Isoflavones and estrogen were introduced into the active site of each platelet receptor; Step 3: Each of the new complexes was taken to the Chimera 1.9 program to obtain the ligand coordinates within the active site.
The grid dimensions were standardized for all bioactive isoflavones and estrogen, using the values 60 × 60 × 60 A, according to the delineation of the respective x, y, z coordinates, with a spacing of 1 Å. Water molecules were removed in Chimera, and the search for multiple ligand conformations within the grid was performed using the Lamarckian Genetic Algorithm (LGA). With the information on the coordinates of the ligands within the active site, molecular anchoring calculations were developed using the Autodock program version 1.5.4 [7].
Using the LigPlot program, only amino acid residues that have interaction between ligands (estradiol and isoflavones) and receptors (enzymes found in the PDB) through Hydrogen or van der Waals bonds are shown in the results.
The physicochemical characteristics that define the degree of affinity and specificity of the ligand for the receptor protein, related to the intermolecular interactions in the receptor/ bioactive complex, were evaluated by hydrogen bonds- Interaction Energy (EI), which refers to the conformation energetically favorable and repulsive interactions between ligand and receptor; and by van der Waals interactions-inhibition constant (Ki), which evaluates the percentage of molecular contact surfaces at each interaction site.
Hydrogen bonds Gibbs interaction energy (EI)-ΔG (Kcal mol-1) are polar intermolecular bonds, where the Hydrogen (H+) of the ligand (bioactive) bonds with the Nitrogen (N¯) of the amino acids of the receivers. It is a strong bond with high intermolecular interaction that shapes the design of the conformational energy of the receptors, determining the conformation of the molecule, and the more negative the E value, the stronger the interaction between the bioactive and the receptor, with platelet G protein-coupled receptors and tyrosine kinases being of interest for research.
van der Waals bonds are forces of attraction or repulsion that act between molecules, and the greater the affinity of the ligand for the receptor, the smaller the distance between the atoms of the molecules. The Interaction Energy (EI) and the interaction inhibition constant (Ki) measured the degree of interaction between the estradiol and isoflavone molecules with the selected platelet receptors.
The assessment of ligand-receptor affinity/acceptance was carried out using the inhibition constant (Ki), with a “bad” affinity/interaction when values in units 10-3 (mM); “good” affinity/interaction with values in units 10-6 (uM); “excellent” affinity/interaction when values in 10-9 units (nM), demonstrating the degree of acceptability between the selected platelet receptors and the molecules being studied in the research.
With the data obtained, the risks of platelet activation for the formation of vascular thrombosis were evaluated, according to the function of receptors coupled to G protein and protein tyrosine kinase, between Estradiol molecules and isoflavone molecules, and between isoflavone molecules themselves. According to Gibbs binding energy (EI) and inhibition constant (Ki).
Validation of the docking calculation between ligands (estradiol and isoflavones) and platelet receptors was carried out by evaluating the binding energy–EI (Kcal mol-1 ), and the inhibition constant–Ki, of the interaction of the receptors with the respective crystallographic ligands.
Result
Platelet receptors from the G protein group and tyrosine kinase enzyme deposited according to their indicated activities, with identification of interaction sites Table 1.
| Receptor /Search | PDB | Action |
| GPVI: G protein and GPVI coupled receptor and collagen and platelets | 2GI7 7NMU 5OU8 | Bounds to collagen and fibrin |
| Stabilizes the clot | ||
| GPIbâ??IXâ??V/PAR: G protein and PAR1-coupled receptor and thrombin and platelets | 1P9A 1OOK GWB 1QYY 1P8V | Main binding site for thrombin |
| P2Y12/ADP: G protein and P2Y12 and ADP coupled receptor and platelets | 4PXZ | Induces platelet response to ADP, increases Ca+ levels in the cytoplasm |
| TP: G protein and TP-coupled receptor and platelets | 6IIU | Recruitment of platelets to the thrombus |
| Cardiovascular homeostasis | ||
| IP (G6b-B): G protein and IP-coupled receptor and prostaglandin and platelets. | 6R0X | Inhibit platelet activation (nitric oxide and prostacyclin), and immunoreceptor tyrosine-based inhibition (ITIM) |
| Bins to heparin and inhibits platelet function | ||
| IP (CLEC2): G protein and IP-coupled receptor and CLEC2 and prostaglandin and PGE2 and platelets | 4YK5 | Catalyzes the formation of PGE2 from PGH2. |
| Cardiovascular risks associated with cyclooxygenase 2 inhibitors (coxibs) | ||
| P2Y1/ADP: G protein and P2Y1 and ADP-coupled receptor and platelets | 4XZ2 | Change in the aggregation form of platelets, |
| Generation of thromboxane A2 | ||
| Adhesion to fibrinogen | ||
| CLEC2: Protein tyrosine kinase and coupled receptor CLEC2 and podoplanin and platelets CLEC2 and platelet and podoplanin | 3IET | Coupled to tyrosine kinase (Syk) |
| Stabilization of the thrombus |
Table 1: Action of platelet receptors.
Table 2 shows the coordinates obtained by Chimera of the active interaction site of each platelet receptor. Of the platelet receptors under study identified in the PDB, only 04 (1P8V; 6IIU; 4YK5; 3IET) presented crystallographic structure of the ligands, Table 3.
| Receptor/Coordinate | X | Y | Z |
| 2GI7 | 4.998 | 14.503 | -21.887 |
| 7NMU | -12.624 | -8.894 | 27.543 |
| 5OU8 | -22.138 | -11.355 | -12.039 |
| 1P9A | -56.304 | 26.415 | -89.947 |
| 1OOK | 18.418 | -16.94 | 117.229 |
| 1GWB | 4.711 | 88.507 | 38.612 |
| 1QYY | -35.196 | -100.561 | -9.753 |
| 1P8V | 35.316 | 38.571 | 36.717 |
| 4PXZ | 16.181 | 1.025 | 59.445 |
| 6IIU | 20.03 | 162.974 | 145.363 |
| 6ROX | 39.917 | -3.762 | 23.337 |
| 4YK5 | 18.402 | 18.725 | 32.539 |
| 4XZ2 | -47.292 | 146.844 | 107.227 |
| 3IET | 51.489 | 13.024 | 10.356 |
Table 2: Values of the coordinates of the active sites of the receivers.
| Rec/Struct. Cryst. Interact. | Hydrogen bonding | van der Waals connection |
| 1P8V/DFP | LEU96 | TYR47, ASN95, TRP92, ARG93, GLU94, ILE179 |
| 6IIU/A8X | HIS89, SER181, ARG295, GLN301 | TRP258, THR298, PHE34, CYS35, MET112, VAL85, ALA31, THR81 |
| 4YK5/4DV | ARG122 | ALA112, VAL108, ILE125, PRO124, GLY109, THR1129, VAL128, LEU132 |
| 3IET/A2G | SER91, ARG98, THR213 | TYR32, VAL97, TRP33, PRO215 |
Table 3: Binding energy and inhibition constant of crystallographed receptors and ligands.
Table 4 shows the amino acid residues of the interaction between each identified receptor and the crystallographic structures of the ligands involved in each study.
| Rec/Struct. Cryst. Interact. | G (Kcal mol-1) | Ki |
| 1P8V/DFP | -4.26 | 756.6 uM |
| 6IIU/A8X | -12.56 | 622.34 pM |
| 4YK5/4DV | -8.6 | 495.23 nM |
| 3IET/A2G | -6.8 | 10.4 uM |
Table 4: Interactions of the receptor and the crystallographic structure and ligand.
GPVI G proteins encoded 2GI7 7NMU 5OU8 bind to the subendothelial collagen of the injured vessel and to fibrin, stabilizing the clot.
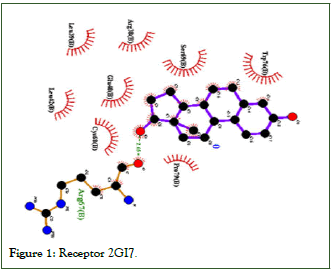
2GI7 receptor Table 5, Estradiol is more selective and has better interaction (EI=-6.01; kcal mol-1; Ki=39.63 uM) that the isoflavones. Among isoflavones, daidzina appeared to be more selective and better interaction, (EI=-5.77; kcal mol-1; Ki=58.78 uM); and glycitein it has lower affinity and lower interaction (EI=-5.11 kcal mol-1; Ki=180.38 uM) in relation to estradiol and other isoflavone molecules. Table 6 shows the amino acid residues by hydrogen bonds and van der Waals bonds of the estradiol and isoflavone ligands. with the receiver. Figure 1 shows the interaction of Estradiol with the platelet receptor 2GI7, thrombotic action with amino acids.
| LINK | E.I | Ki |
| Estradiol | -6, 01 | 39,63 (uM) |
| Daidzeina | -5,26 | 139,71 (uM) |
| Daidzina | -5,77 | 58,78 (uM) |
| Genistein | -5,19 | 157,55 (uM) |
| Genistin | -5,63 | 74,04 (uM) |
| Glycytein | -5,11 | 180,38 (uM) |
| Glycitin | -5,57 | 82,59 (uM) |
| Note: E.I: Interact Energy (kcal mol-1); Ki=Inhibition Constant | ||
Table 5: Receptor 2GI7.
| Complex 2GI7 | Hydrogen bonding | van der Waals |
| Estradiol | ARG67 | TRP76, SER69, ARG38, GLU40, PRO79, CYS68, LEU42, LEU39 |
| Daidzeina | ARG38, SER69, GLU40, ARG67 | TRP76, SER77, PRO79, CYS68, LEU39 |
| Daidzina | ASP49, GLN50, TYR47, ALA57 | GLN48, AGN46, LEU53, ILE55, PRO56, LEU62 |
| Genistein | TYR47, ASP49, SER45 | GLN48, ARG46 |
| Genistin | ARG38, SER69, GLU40, ARG67, GLN82 | CYS68, PRO79, ARG65, LEU42 |
| Glycytein | ARG67, ARG38 | LEU42, GLU40, SER69, SER77, TRP76, LEU39, CYS68 |
| Glycitin | ARG65, GLN82, ARG67, SER69, ARG38 | LEU42, GLU40, PRO79, CYS68 |
Table 6: Hydrogen bonding and van der Waals interactions in complex 2GI7.
Figure 1: Receptor 2GI7.
The interaction of Estradiol with the platelet receptor 2GI7 occurs through hydrogen bonds with arginine (ARG67) and through van der Waals bonds with tryptophan (TRP76), serine (SER69), arginine (ARG38), glutamate (GLU40), proline (PRO79), cysteine (CYS68), leucine (LEU39.
7NMU receiver Table 7, both estradiol and isoflavones had good affinity, however the Glycitin is more selective and has better interaction (EI=-7.17; kcal mol-1; Ki=5.58 uM one). Glycitein showed lower affinity for the receptor ((EI=-5.63 kcal mol-1; Ki=74.21 uM) for the platelet receptor 7NMU. Table 8 shows the amino acid residues by hydrogen bonds and van der Waals bonds of the estradiol and isoflavones with the receiver.
| LINK | E.I | Ki |
| Estradiol | -6.72 | 11.88 uM |
| Daidzeina | -6.1 | 34.04 uM |
| Daidzina | -7.14 | 5.83 uM |
| Genistein | -6.16 | 30.66 uM |
| Genistin | -6.84 | 9.64 uM |
| Glycytein | -5.63 | 74.21 uM |
| Glycitin | -7.17 | 5.58 uM |
| Note: EI: Interact Energy (kcal mol-1); Ki: Inhibition Constant | ||
Table 7: Receptor 7MNU.
| Complex 7NMU | Hydrogen bonding | van der Waals |
| Estradiol | GLU6, TRP116 | GLN5, GLN3, LEU4, VAL2, GLN118, GLY117 |
| Daidzeina | GLU6, LEU4 | GLN118, GLY117, TRP116, TYR115, VAL2, GLN3 |
| Daidzina | GLN5, GLU6, GLN118 | GLY117, LEU4, GLN3, TRP116, TYR115, VAL2 |
| Genistein | LEU4, GLU6 | GLN118, TRP116, GLY117, TYR115, GLN2, VAL2 |
| Genistin | GLU6, ASP114 | GLN5, GLN118, LEU4, TYR115, GLN3, VAL2, TRP116 |
| Glycytein | LEU4, ASP114 | GLN5, GLU6, GLY117, GLN118, TRP116, TYR115, GLN3, VAL2 |
| Glycitin | GLN5, GLU6, GLN118 | GLY117, GLN3, LEU4, TYR115, VAL2, TRP116 |
Table 8: Hydrogen bonding and van der Waals interactions in complex 7NMU.
For the 5OU8 receiver Table 9, the Genistin is more selective and has better interaction (EI=-6.64; kcal mol-1; Ki=13.55 uM) with the receptors that Estradiol (EI=-6.05; kcal mol-1; Ki=37.01 uM) and that the other isoflavones , and genistein presents lower affinity and lower interaction (EI=-5.73 kcal mol-1 ; Ki=63 uM). Table 10 shows the amino acid residues by hydrogen bonds and van der Waals bonds of the estradiol and isoflavone ligands with the receiver.
| LINK | E.I | Ki |
| Estradiol | -6.05 | 37.01 uM |
| Daidzeina | -6.02 | 38.73 uM |
| Daidzina | -6.57 | 15.17 uM |
| Genistein | -5.73 | 63.6 uM |
| Genistin | -6.64 | 13.55 uM |
| Glycytein | -6.03 | 38.13 uM |
| Glycitin | -6.37 | 21.51 uM |
| Note: E.I=Interact. Energy (kcal mol-1). Ki=Inhibition Constant | ||
Table 9: Receptor 5OU8.
| Complex 5OU8 | Hydrogen bonding | van der Waals |
| Estradiol | GLU40, ARG38, | TRP76, SER77, PRO79, LEU78, ARG67, SER69 |
| Daidzeina | ARG38, GLU40, SER69 | TRP76, PRO79, LEU78, SER77 |
| Daidzina | ARG67, SER69, SER77, GLN1 | PRO79, LEU78, TRP76, LEU5, LEU75, PRO4, GLY3 |
| Genistein | GLU40, ARG38, SER69 | TRP76, PRO79, SER77, LEU78 |
| Genistin | GLN1, LEU5, TRP76, SER77 | PRO79, LEU78, PRO4, GLY3, LEU75 |
| Glycytein | ARG38, GLU40, SER69 | LEU78, PRO79, SER77, TRP76 |
| Glycitin | GLN1, AER69, SER77, ARG67 | LEU75, LEU5, LEU78, TRP76, GLY3, PRO4, PRO79 |
Table 10: Hydrogen bonding and van der Waals interactions in complex 5OU8.
1P9A receiver Table 11 shows that Daidzina (EI=-6.56; Ki=15.63 uM ) is more selective and has better interaction with the receptor among the ligands evaluated, and Estradiol is less selective than isoflavones for this receptor (EI=-5.78 kcal mol-1; Ki=58.38 uM). Table 12 shows the amino acid residues by hydrogen bonds and van der Waals bonds of the estradiol and isoflavone ligands with the receiver.
| LINK | E.I | Ki |
| Estradiol | -5,78 | 58,38 (uM) |
| Daidzeina | -6,4 | 20,46 (uM) |
| Daidzina | -6,56 | 15,63 (uM) |
| Genistein | -6,09 | 34,49 (uM) |
| Genistin | -6,11 | 33,19 (uM) |
| Glycytein | -6,18 | 29,62 (uM) |
| Glycitin | -6,17 | 30,14 (uM) |
Table 11: Receptor 5OU8.
| Complex 1P9A | Hydrogen bonding | van der Waals |
| Estradiol | GLU151, LEU196 | ASN173, GLU172, HIS195, PRO198, ASP175, LEU174 |
| Daidzeina | LYS152, THR176, VAL236, ASP235 | LYS237, PHE199, LEU178, TYR130 |
| Daidzina | LYS152, ASN173, HIS195, GLU172, ASP235, VAL236 | ASP175, THR176, PHE199 |
| Genistein | LYS237, VAL236, THR176, LYS152 | PHE199, ASP235, TYR130, LEU178 |
| Genistin | ASP235, LYS152, ASN173, LEU196, HIS195, GLU172 | ASP175, PRO198, PHE199, VAL236 |
| Glycytein | VAL236, THR176, LYS152, ASP235 | TRP230, TYR130, LEU178, LYS237, PHE199 |
| Glycitin | ASN173, HIS195, LYS152, GLU172, ASP235, VAL236 | ASP175, PHE199, GLU151, THR176 |
Table 12: Hydrogen bonding and van der Waals interactions in complex 1P9A.
1OOK receiver Table 13, a glycitin it is the most selective and has the best interaction (EI=-6.93 kcal mol-1; Ki=8.39 uM). Daidzein has lower affinity and smaller interaction (EI=-5.71 kcal mol-1; Ki=65.1 uM) with the receptor.
| LINK | E.I | Ki |
| Estradiol | -6,68 | 12,78 (uM) |
| Daidzeina | -5,71 | 65,1 (uM) |
| Daidzina | -6,9 | 8,75 (uM) |
| Genistein | -5,83 | 53,35 (uM) |
| Genistin | -6,82 | 9,94 (uM) |
| Glycytein | -5,74 | 62,09 (uM) |
| Glycitin | -6,93 | 8,39 (uM) |
| ÃÂ? Note: E.I=Interact. Energy (kcal mol-1). Ki=Inhibition Constant | ||
Table 13: Receptor 1OOK.
Table 14 shows the amino acid residues by hydrogen bonds and van der Waals bonds of the estradiol and isoflavone ligands with the receiver.
| Complex 1OOK | Hydrogen bonding | van der Waals |
| Estradiol | LYS60F, ARG3, ASN143 | HIS57, GLY193, GLU192, ARG73, GLN151, LEU40, LEU41 |
| Daidzeina | LYS60F, ASN143 | HIS57, LEU41, TRP60D, ARG3, GLY193, GLU192, ARG73, GLN151 |
| Daidzina | ASN143, GLY142, ARG73, LYS60F, TRP60D | TRP141, GLY193, LEU41, LEU40, PHE60H, ASP60E |
| Genistein | LYS60F, ASN143 | GLN151, ARG73, LEU40, LEU41, GLY193, GLU192, ARG3, HIS57, TEP60D |
| Genistin | ARG73, GLU39, ARG3, LYS60F | TRP60D, HIS57, LEU40, LEU41, GLY193, GLN38 |
| Glycytein | PRO2, LYS60F, THR147 | TRP148, TRP60D, HIS57, CYS42, GLY193, ARG3, GLU192 |
| Glycitin | ASN143, LYS60F, ARG173, TRP141 | GLY142, LEU40, LEU41, ARG73, PHE60H, ARG3, GLY193, TRP60D |
Table 14: Hydrogen bonding and van der Waals interactions in complex 1OOK.
1GWB receiver Table 15 a daidzina it is more selective and has better interaction (-7.29 kcal mol-1; Ki=4.55 uM), compared to estradiol (EI=-7.29 kcal mol-1; Ki=9.99 uM) and the other isoflavones. And among the ligands, daidzein showed lower affinity and smaller interaction (EI=-6.43 kcal mol-1; Ki=19.48 uM). Table 16 shows the amino acid residues by hydrogen bonds and van der Waals bonds of the estradiol and isoflavone ligands with the receiver.
| LINK | E.I | Ki |
| Estradiol | -6,82 | 9,99 (uM) |
| Daidzeina | -6,43 | 19,48 (uM) |
| Daidzina | -7,29 | 4,55 (uM) |
| Genistein | -6,76 | 11,0 (uM) |
| Genistin | -7,07 | 6,53 (uM) |
| Glycytein | -6,5 | 17,34 (uM) |
| Glycitin | -7,17 | 5,59 (uM) |
| Note: E.I=Interact. Energy (kcal mol-1), Ki=Inhibition Constant | ||
Table 15: Receptor 1GWB.
| Complex 1GWB | Hydrogen bonding | van der Waals |
| Estradiol | ASN221, GLY220 | GLN263, CYS264, LYS269, ASP268, ASP265, SER267 |
| Daidzeina | LYS269, ASP268, ASN221 | GLY220, ASP265, CYS264, GLN263, SER267, |
| Daidzina | LYS269, ASN221, ASP268 | GLN263, CYS264, ASP265, SER267 |
| Genistein | GLY220, SER261, CYS264, ASP268 | VAL262, HIS219, GLN263, LYS269, SER267, ASN221, ASP265 |
| Genistin | ASP268, CYS264, ASP265, GLY220 | SER267, LYS269, GLN263, VAL262, SER261, ASN221, HIS219 |
| Glycytein | ASN221, GLY220 | TRP223, SER261, GLN263, ASP265, LYS269, ASP268 |
| Glycitin | SER267, ASP268, LYS269, ASN221 | GLN263, CYS264, ASP265 |
Table 16: Hydrogen bonding and van der Waals interactions in complex 1GWB.
1QYY Receiver Table 17 shows that the Daidzina (EI=-6.32; kcal mol-1; Ki=23.3 uM) is more selective and has better interaction with platelet G protein receptors 1QYY compared to Estradiol (EI=-6.27 kcal mol-1; Ki=25.49 uM). Glycitein (EI=-5.5 kcal mol-1; Ki=92.3 uM) showed lower affinity and less interaction with the receptor. Table 18 shows the amino acid residues by hydrogen bonds and van der Waals bonds of the estradiol and isoflavone ligands with the receiver.
| LINK | E.I | Ki |
| Estradiol | -6,27 | 25,49 (uM) |
| Daidzeina | -5,51 | 91,72 (uM) |
| Daidzina | -6,32 | 23,3 (uM) |
| Genistein | -5,56 | 84,08 (uM) |
| Genistin | -5,88 | 48,92 (uM) |
| Glycytein | -5,5 | 92,3 (uM) |
| Glycitin | -5,92 | 45,78(uM) |
| Note: E.I=Interact. Energy (kcal mol-1), Ki=Inhibition Constant | ||
Table 17: Receptor 1QYY.
| Complex 1QYY | Hydrogen bonding | van der Waals |
| Estradiol | ASN223 | ARG218, ASP222, TRP219, SER194, GLY193, TYR215 |
| Daidzeina | HIS195 | GLY193, TRP219, LEU196, TYR215, ARG218, ASP222, ASN223 |
| Daidzina | TRP219 | HIS195, GLY193, PHE192, ASN223, TYR215, ARG218, THR266 |
| Genistein | TYR215, ASN223 | ASP222, ARG218, TRP219, SER194, LEU196, HIS195 |
| Genistin | SER194, ASP222, ASP223 | ARG218, TRP219, TYR215, GLY193, LEU196 |
| Glycytein | LEU196 | HIS195, GLY193, TRP219, ASN223, ASP222, ASG218, TYR215, |
| Glycitin | ASP222, GLN221, LYS258 | TYR259, ALA224, TYR257, PRO260 |
Table 18: Hydrogen bonding and van der Waals interactions in complex 1QYY.
1P8V platelet receiver Table 19 , shows that between the isoflavones to Daidzein (EI=-8.5 kcal mol-1 ; Ki=591.65 nM), to glycitin (EI=-8.32 kcal mol-1 ; Ki=795.17 nM) and Estradiol (EI=-8.31 kcal mol-1; Ki=812.13 nM) present higher selectivity and greater affinity. Daidzina appears to be less selective and with lower affinity (EI=-6.84 kcal mol-1; Ki=9.7 uM). Redocking to evaluate the interactions of the ligands under study with the 1P8V receptor was carried out using the crystallographic structure of the DFP ligand complex (Diisopropyl Fluorophosphate) which is an inactivated human thrombin, with the GPIb α subunit of the GPIb-IX-V complex (1P8V), presenting a less favorable Gibbs energy than the ligands studied. For the crystallographed ligand, the value obtained was EI=-4.26 kcal mol-1; Ki=756.6 nM. Table 20 shows the amino acid residues by hydrogen bonds and van der Waals bonds of the estradiol and isoflavone ligands with the receiver. The DPF ligand forms a hydrogen bond with LEU96, and van der Waals interactions with residues of TYR47, ASN95, TRP92, ARG93, GLU94, ILE179. Only the amino acids TYR47 and GLU94 coincide with the linker amino acids studied–Estradiol and Is flavones.
| LINK | E.I | Ki |
| Estradiol | -8,31 | 812,13 (nM) |
| Daidzeina | -8,5 | 591,65 (nM) |
| Daidzina | -6,84 | 9,7 (uM) |
| Genistein | -6,88 | 9,01 (uM) |
| Genistin | -8,05 | 1,25 (uM) |
| Glycytein | -6,84 | 9,64 (uM) |
| Glycitin | -8,32 | 795,17 (nM) |
| DFP | - 4,26 | Â 756,6 (uM) |
| Note: E.I=Interact. Energy (kcal mol-1), Ki=Inhibition Constant | ||
Table 19: Receptor 1P8V.
| Complex 1p8v | Hydrogen bonding | van der Waals |
| Estradiol | GLU202, TYR240 | ASP199, ALA200, CYS231, GLY230, VAL225, SER226, GLY238, PHE239, TRP227 |
| Daidzeina | GLY228, TRP92, GLY230, TRP227 | CYS231, VAL225, SER226, HIS43, LEU96, TYR47, PHE239, ALA200, A//SP199, CYS231 |
| Daidzina | ASP199 | ALA200, GLY238, TRP227, VAL225, CYS201, GLU202, GLY203, SER205, HIS43 |
| Genistein | SER205, HIS43, LYS52 | CYS201, GLY230, GLY228, TRP50, TYR47 |
| Genistin | GLY203, GLY230, CYS44 | PHE54, LYS52, HIS43, LEU27, GLU202, SER205, CYS28, VAL225, ALA200, ASP199, CYS231, TRP227, PHE239, CYS231 |
| Glycytein | LYS52, GLN156, ASN143 | GLU202, GLY142, TRP141, GLY203, LEU26, LEU27, CYS28, CYS44 |
| Glycitin | GLY230, SER205, GLY228, SER226 | CYS201, ALA200, ASP199, PHE239, TRP227, VAL225, HIS43, LEU96, TYR47, GLU94, CYS231 |
Table 20: Hydrogen bonding and van der Waals interactions in complex 1p8v.
For 4PXZ receiver (P2Y12/ADP) Table 21, shows that the Daidzina (EI=-8.41; kcal mol-1; Ki =688.93 nM) and the glycitin (EI=-8.21 kcal mol-1; Ki=951.38 nM) present greater selectivity and affinity with the indicative receptor in relation to the other ligands. The Estradiol and Daidzein molecules, with the same values of the inhibition constant (Ki=1.89 uM), show similar interaction for the 4PXZ receptor, with lower affinity for the protein-coupled platelet receptor G P2Y12 PDB-4PXZ) in relation to the other ligands. Table 22 shows the amino acid residues by hydrogen bonds and van der Waals bonds of the estradiol and isoflavone ligands with the receiver.
| LINK | E.I | Ki |
| Estradiol | -7.81 | 1.89 uM |
| Daidzeina | -7.81 | 1.89 uM |
| Daidzina | -8.41 | 688.93 nM |
| Genistein | -7.22 | 5.08 uM |
| Genistin | -7.7 | 2.27 uM |
| Glycytein | -7.66 | 2.44 uM |
| Glycitin | -8.21 | 951.38 nM |
| Note: E.I: Interact. Energy (kcal mol-1), Ki=Inhibition Constant | ||
Table 21: Receptor 4PXZ.
| Complex 1P8V | Hydrogen bonding | van der Waals |
| Estradiol | CYS97 | CYS194, ASN191, TYR109, VAL190, ASN159, ARG256, TYR105, SER101, CYS175, HIS187, LYS179, VAL102 |
| Daidzeina | CYS97, TRY109, ASN191 | ASN159, LYS179, HIS187, ARG256, VAL190, VAL102, TRY105, CYS194 |
| Daidzina | ASN159, SER101, CYS194, ASN191, ARG256 | VAL102, SER156, TYR105, TYR109, VAL190, CYS175, HIS187, TYR259, LYS174, PHE277, GLN263, LYS280, ARG93, GLU281 |
| Genistein | CYS97, TYR109 | LYS179, ASN159, ARG256, HIS187, ASN191, TYR105, CYS194, VAL102, VAL190 |
| Genistin | CYS97 | CYS194, ASN191, TYR109, VAL190, ASN159, ARG256, TYR105, SER101, CYS175, HIS187, LYS179, VAL102 |
| Glycytein | CYS97, LYS179, ASN191 | VAL102, CYS175, HIS187, ASN159, VAL190, TYR105, CYS194, TYR109, ARG256 |
| Glycitin | ASN159, CYS194, ASN191, SER101, ARG256 | SER156, VAL102, HIS187, VAL190, TYR109, TYR105, LYS280, CYS175, TYR259, GLU281, GLN263, PH3277, LYS174, ARG93 |
Table 22: Hydrogen bonding and van der Waals interactions in complex 1p8v.
For the 6IIU receiver Table 23 shows that The Daidzina (EI=-8.49 kcal mol-1; Ki=598.79 nM) has excellent selectivity and affinity for the platelet G protein receptor TP. Redocking to evaluate the interactions of the ligands under study with the receptor was carried out using the crystallographic structure of the A8X–Ramatroban ligand complex , which presented a more favorable interaction energy than the molecules studied (EI=-12.56 kcal mol-1; Ki=622.34 pM). It is important to highlight that the A8X ligand is a thromboxane–TP receptor blocker, used as an inhibitor of cardiovascular dysfunction and not in HRT in menopause. A Daidzein (EI=-6.97; kcal mol-1; Ki=7.78 uM) has lower selectivity and lower affinity for this receiver. Table 24 shows the amino acid residues by hydrogen bonds and van der Waals bonds of the estradiol and isoflavone ligands with the receiver. The A8X ligand makes hydrogen bonds with HIS89, SER181, ARG295, GLN301, and van der Waals interactions with residues of TYR47, ASN95, TRP92, ARG93, GLU94, ILE179. Except for the amino acids PHE34 and CYS35, all the other amino acids coincide with the linker amino acids studied.
| LINK | E.I | Ki |
| Estradiol | -8.16 | 1.04 uM |
| Daidzeina | -6.97 | 7.78 uM |
| Daidzina | -8.49 | 598.79 nM |
| Genistein | -7.09 | 6.4 uM |
| Genistin | -8.13 | 1.09 uM |
| Glycytein | -7.72 | 2.21 uM |
| Glycitin | -7.59 | 2.73 uM |
| A8X | -12.56 | 622.34 pM |
Table 23: Receptor 6IIU.
| Complex 6IIU | Hydrogen bonding | van der Waals |
| Estradiol | ARG295, THR81 | MET112, THR298, LEU78, VAL85, SER181, PRO179, LEU291, LEU294, HIS89 |
| Daidzeina | THR298, HIS89, SER181, GLN301 | MET112, THR81, TRP182, LEU294 ARG295, VAL85, PRO179 |
| Daidzina | MET112, THR298, LEU294, THR81, CYS35 | PHE115, GLY116, TRP258, LEU78, ALA297, ALA31, VAL85, PHE30, HIS89 |
| Genistein | PHE115, THR298, LEU294, SER181, HIS89 | ALA297, GLN301, MET112, VAL85, THR81, TRP182 |
| Genistin | MET112, THR298, LEU294, CYS35 | GLY116, TRP258, ALA297, PHE115, GLY82, ALA31, LEU78, VAL85, ARG295, PHE30, HIS89 |
| Glycytein | GLNE301, THR298, ARG295, HIS89, SER181 | THR81, MET112, LEU294, PRO179, VAL85, TRP182 |
| Glycitin | MET112, LEU294, THR298, GLN301 | PHE200, GLY116, TRP258, LEU78, THR81, HIS89, VAL85, ARG295, ALA31, LEU261, PHE184, ALA297 |
Table 24: Hydrogen bonding and van der Waals interactions in complex 6IIU.
The platelet receptor 6R0X (IP (G6b-B)) Table 25 shows that Estradiol (EI=-5.7 kcal mol-1; Ki=66.52 uM) has greater selectivity and affinity for the receptor, indicating a lower risk of thrombus formation as described on the receptor function. And among the molecules of isoflavones a Daidzina (EI=-5.3; kcal mol-1; Ki=130.75 uM) has greater selectivity and affinity for the receptor with lower risk of thrombosis, and Genistin is less selective and with lower affinity (EI=-4.71 kcal mol-1; Ki=352.15 uM) with the receptor revealing no protection against the risk of thrombosis in this interaction.
| LINK | E.I | Ki |
| Estradiol | -5.7 | 66.52 uM |
| Daidzeina | -5.11 | 180.74 uM |
| Daidzina | -5.3 | 130.75 uM |
| Genistein | -4.78 | 312.76 uM |
| Genistin | -4.71 | 352.15 uM |
| Glycytein | -4.96 | 231.27 uM |
| Glycitin | -4.73 | 341.47 uM |
| Note: E.I: Interact. Energy (kcal mol-1); Ki: Inhibition Constant | ||
Table 25: Receptor 6IIU.
Table 26 shows the amino acid residues by hydrogen bonds and van der Waals bonds of the estradiol and isoflavone ligands with the receiver. Figure 2 shows the interaction of Estradiol with the platelet receptor 6ROX, antithrombotic action with amino acids.
| Complex 6R0X | Hydrogen bonding | van der Waals |
| Estradiol | GLY77, VAL78, THR73 | HIS75, LEU74 |
| Daidzeina | SER76, GLY77 | LEU74, HIS75, SER80, VAL78, PRO79 |
| Daidzina | GLY77, LEU74, THR73, SER72, TYR70, ASN51 | HIS75, SER76 |
| Genistein | SER76, GLY77 | HIS75, LEU74, SER80, PRO79, VAL78 |
| Genistin | GLY77, HIS75, VAL78, LEU74 | SER76, THR73, TYR70, SER72 |
| Glycytein | SER72 | SER76, GLY77, HIS75, VAL78, THR73, LEU74 |
| Glycitin | SER76, LEU74 | THR73, TYR69, HIS75, GLY77, VAL78, SER80, PRO79 |
Table 26: Hydrogen bonding and van der Waals interactions in complex 6R0X.
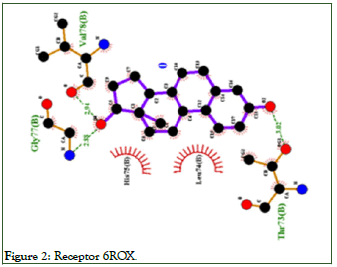
Figure 2: Receptor 6ROX.
The interaction of Estradiol with the platelet receptor 6ROX occurs through hydrogen bonds with the amino acids GLY77, VAL78, THR73, and through van der Waals bonds with the amino acids HIS75, LEU74.
4YK5 receptor (IP (CLEC2)-mPGES-1 inhibitory complex Table 27 shows that Estradiol (EI=-4.53 kcal mol-1; Ki=481.93 uM) compared to isoflavones presents greater selectivity and greater affinity. The isoflavone molecules under study present an unfavorable interaction (values in mM) with this receptor. A genistin is the isoflavone with the lowest affinity for the receptor (Ki=2.67 mM) showing lower cardiovascular risk. Redocking to evaluate the ligands under study was carried out using the crystallographed structure with 4DV (Dimethyl Propanoic Acid). The redocking the 4DV ligand has a more favorable interaction energy (EI=-8.6 kcal mol-1, Ki=495.23 nM) than the molecules studied (estradiol and isoflavones). Table 28 shows the amino acid residues by hydrogen bonds and van der Waals bonds of the estradiol and isoflavone ligands with the receiver. The 4DV ligand forms a hydrogen bond with ARG122, and van der Waals interactions with residues ALA112, VAL108, ILE125, PRO124, GLY109, THR129, VAL128, LEU132. Except for the amino acids VAL108, GLY109 and LEU132, all the others amino acids coincide with the amino acids from estradiol and isoflavones studied. Glycitein did not interact via hydrogen bonds with the aforementioned receptor.
| LINK | E.I | Ki |
| Estradiol | -4.53 | 481.93 uM |
| Daidzeina | -3.99 | 1.19 mM |
| Daidzina | -3.88 | 1.44 mM |
| Genistein | -3.91 | 1.35 mM |
| Genistin | -3.51 | 2.67 mM |
| Glycytein | -3.95 | 1.28 mM |
| Glycitin | -3.61 | 2.24 mM |
| 4DV | -8.6 | 495.23 nM |
| Note: EI: Interact. Energy (kcal mol-1); Ki: Inhibition Constant | ||
Table 27: Receptor 4YK5.
| Complex 4YK5 | Hydrogen bonding | van der Waals |
| Estradiol | ARG122 | THR129, ILE125, VAL128, PRO124 |
| Daidzeina | ARG126, LEU121 | ILE125, ALA123, SER127, PRO124, ARG122 |
| Daidzina | ALA123, ARG122 | ALA112, THR129, VAL128, ILE125, PRO124 |
| Genistein | ALA123, ILE125 | THR129, VAL128, PRO124, ARG122 |
| Genistin | ARG122, ALA123 | ALA112, THR129, ILE125, VAL128, PRO124 |
| Glycytein | â?? | VAL128, THR129, ILE125, ALA123, PRO124, ARG122 |
| Glycitin | ALA123, ARG122 | THR129, ILE125, VAL128, PRO124 |
Table 28: Hydrogen bonding and van der Waals interactions in complex 4YK5.
For the platelet receptor 4XZ2 (Tyrosine kinase P2Y1/ATP) Table 29, shows that Estradiol (EI=-8.26 kcal mol-1; Ki=888.57 nM) has greater selectivity and affinity. Among the isoflavone molecules, Daidzein (EI=-6.65 kcal mol-1; Ki=13.44 uM) it presents lower selectivity and lower affinity. Table 30 shows the amino acid residues by hydrogen bonds and van der Waals bonds of the ligands estradiol and isoflavones with the receiver.
| LINK | E.I | Ki |
| Estradiol | -8.26 | 888.57 nM |
| Daidzeina | -6.65 | 13.44 uM |
| Daidzina | -7.11 | 6.12 uM |
| Genistein | -7.06 | 6.67 uM |
| Genistin | -6.93 | 8.38 uM |
| Glycytein | -6.93 | 8.38 uM |
| Glycitin | -6.91 | 8.56 uM |
| Note: EI: Interact. Energy (kcal mol-1), Ki: Inhibition Constant | ||
Table 29: Receptor 4XZ2.
| Complex 4XZ2 | Hydrogen bonding | van der Waals |
| Estradiol | CYS98, ALA96 | LEU136, ARG107, ARG102, PHE101, GLY129, SER32, TYR64, ARG97 |
| Daidzeina | ASP128 | PHE101, TYR64, CYS98, ALA96, ARG102, SER32, SER130, GLY33, GLY127, ARG97, GLY129 |
| Daidzina | ARG383, ASP177, HIS220, ARG219, ASP128 | THR364, CYS221, GLN368, GLY129, THR132, GLY133, LEU136 |
| Genistein | ASP128, SER130, GLY127, CYS98 | PHE101, TYR64, ALA96, SER32, GLY33, ARG102, ARG97, GLY129, ARG219 |
| Genistin | ARG383, ASP177, HIS220, ARG219 | GLN365, GLN368, THR364, VAL367, THR132, ASP128, CYS221 |
| Glycytein | ASP128 | ARG102, CYS98, GLY129, SER130, GLY127, ALA96, PHE101, TYR64, GLY33, ARG97, SER32 |
| Glycitin | ARG383, ASP177, HIS220, ARG219, ASP128 | GLY133, THR132, GLY129, GLN368, THR364, VAL367, CYS221 |
Table 30: Hydrogen bonding and van der Waals interactions in complex 4XZ2.
3IET receptor (Tyrosine kinase Syk (CLEC2)) Table 31 shows that Daidzina has greater selectivity and the highest affinity (EI=-8.29 kcal mol-1; Ki=840.49 nM). Genistein (EI=-6.7 kcal mol-1; Ki=12.31 uM), although it has good interaction with the receptor, presents the lowest selectivity and lowest affinity among isoflavone molecules. Redocking to evaluate the interactions of the ligands under study with the 3IET receptor was carried out using the crystallographed structure of the A 2G ligand complex, which showed interaction energy EI=-6.8 kcal mol-1, Ki=10.4 uM equivalent to the ligand molecules in this research. Table 32 shows the amino acid residues by hydrogen bonds and van der Waals bonds of the estradiol and isoflavone ligands with the receptor.
| LINK | E.I | Ki |
| Estradiol | -7.91 | 1.59 uM |
| Daidzeina | -7.44 | 3.5 uM |
| Daidzina | -8.29 | 840.49 nM |
| Genistein | -6.7 | 12.31 uM |
| Genistin | -8.02 | 1.33 uM |
| Glycytein | -6.98 | 7.71 uM |
| Glycitin | -7.4 | 3.74 uM |
| A2G | -6.8 | 10.4 uM |
| Note: E.I: Interact Energy (kcal mol-1); Ki: Inhibition Constant | ||
Table 31: Receptor 3IET.
| Complex 3EIT | Hydrogen bonding | van der Waals |
| Estradiol | GLU61, GLU50, ARG52 | VAL94, TRY59, THR213, TYR58, LYS214, TRP47, PRO95 |
| Daidzeina | ARG98, SER91 | ASN99, HIS34, TYR49, VAL97, TYR36, TRP33, PRO215, TYR32, PRO216 |
| Daidzina | ARG52, GLU61, LYS64, TYR59, | THR92, TYR32, SER91, PRO215, TRP33, THR213, LYS214, ARG98, PRO95, HIS93, TYR58, VL94, GLU50, TRP47, ALA60 |
| Genistein | TYR59, SER91 | PRO95, TRP47, LYS64, TYR58, ARG52, THR213, HIS93, TRP33, ARG98, GLU50, VAL94 |
| Genistin | ARG52, TYR59, LYS64, GLU61 | TYR32, THR92, PRO215, TRP33, SER91, HIS27D, ARG98, GLU50, VAL94, HIS93, PRO95, TYR58, THR213, LYS214, ALA60, TRP47 |
| Glycytein | LYS64, TYR59, SER91 | HIS93, TRP33, THR213, ARG98, GLU50, PRO95, TYR58, TRP47 ARG52, VAL94 |
| Glycitin | TYR59, THR57, SER91 | ARG98, TRP47, HIS93, ARG52, TYR58, LYS64, TRP33, PRO95, THR213, GLU50, VAL94, ALA60, GLU61 |
| A2G | SER91, ARG98, THR213 | TYR32, VAL97, TRP33, PRO215 |
Table 32: Hydrogen bonding and van der Waals interactions in complex 3EIT.
The ligand A2G–N-acetyl-α-D-galactosamine, is a carbohydrate derivative, involved in the formation of glycoproteins; antigenic action and cellular interaction action, but it is not a compound for use in menopausal symptoms. Interacts with the 3IET receiver through hydrogen bonding with SER91, ARG98, THR213, and through van der Waals bonding with residues of TYR32, VAL97, TRP33, PRO215. Except amino acids ARG98 and THR213, all the others amino acids coincide with the linker amino acids studied.
Discussion
It is known that women are generally protected from cardiovascular diseases before menopause, but after menopause they are at greater risk compared to men. Studies have indicated that postmenopausal women have higher platelet activity than premenopausal women, which may increase the risk of thromboembolic events and cardiovascular diseases. The literature differs regarding the prothrombotic effects of estrogen replacement therapy, regarding the increase or decrease in platelet activation and adhesion due to the inhibition of the expression of the platelet receptor GP IIb/IIIa, justifying further investigation.
There is a consensus in the literature that the G protein-coupled receptor GPVI is an important receptor for contributing to the appearance of thrombosis triggered by exposed subendothelial collagen with platelet aggregation action, which, by interacting with fibrin, promotes clot stabilization. It has been identified as a physiological collagen receptor and as a promising target for antithrombotic drugs. And although Feitsma et al report what Essential details of the interaction of the GPVI receptor with subendothelial collagen remain elusive, states that Glycoprotein VI (GPVI) mediates collagen-induced platelet activation through the essential amino acid residues TRP76, ARG38 AND GLU40. In our study, a good interaction between Estradiol and isoflavones with this receptor was demonstrated, with less intensity of interaction between isoflavones. Glycytein Genistein Glycitein.
Estradiol-induced platelet activation for the platelet GPVI receptor PDB-2GI7 is mediated mainly through interaction with the amino acid residue ARG67; for the PDB-7NMU receptor, the interaction is mediated mainly by the amino acid residues GLU6 and TRP116; it is to the GPVI receptor PDB-5OU8 by the amino acid residues GLU40 and ARG38. We can conclude that for Estradiol, the platelet receptor coupled to the GPVI G protein, the interaction with the amino acid residues of glutamate and arginine predominates, with better interaction with glutamate (EI=-2.55 kcal mol-1).
The interaction of bioactive molecules with platelet receptors PAR (Receptors Activated by Proteases) were encoded as platelet receptors PDB-1P9A, PDB-1OOK, PDB-1GWB, PDB-1QYY PDB-1P8V. These receptors are the main binding sites for thrombin. Among them, the one with the best interaction with the bioactives studied, estradiol and isoflavones, was the G protein- coupled receptor PDB-1P8V. And among the molecules tested, Estradiol showed the best interaction for this receptor followed by Glycitin and Daidzein.
Estradiol-induced platelet activation for the platelet receptor PAR PDB-1P8V is mediated mainly through interaction with the amino acid residue GLU202, TYR240. Glycitin interacts with the PDB-1P8V receptor mainly by the amino acid residues GLY230, GLY228, SER226, SER205.
According to the results of the interaction of each ligand with the receptor of the receptor platelet P2Y12 PDB-4PXZ, which is activated by ADP, elevating Ca+ levels in the platelet cytoplasm, demonstrated that Daidzein and Glycitin had a strong interaction with the receptor, and even greater than the interaction of this receptor with Estradiol. It was identified that Estradiol-induced platelet activation for this platelet receptor is mediated mainly through interaction with the amino acid residue CYS97.
As previously reported, the literature describes that the TP PDB-6IIU receptor, when activated, induces the recruitment of platelets to the thrombus and promotes vasoconstriction in vascular smooth muscle, which were identified in the placenta and in human umbilical vein endothelial cells. Thromboxane in the placenta is produced by trophoblast cells and prostacyclin by endothelial cells. Thus, in clinical practice, as aspirin is used in low doses to block platelet TXA2 synthesis, it crosses the placenta and its levels decrease, sparing the inhibition of prostacyclin. However, aspirin can cause gastrointestinal bleeding and hemorrhagic stroke, and there is still a significant percentage of patients who do not respond to aspirin, as aspirin is unable to inhibit the formation of isoprostanes. which are platelet TP receptor agonists and can partially activate the TXA signaling pathway. Therefore, TP receptor antagonists highly selective and potent drugs would still be of great clinical interest for cardiovascular diseases aiming at interaction with the PDB-6IIU TP receptor, benefiting the rational design of more selective and high affinity drugs as therapeutic agents. Aiming at hormone replacement in menopause, our study for the platelet receptor TP PDB-6IIU revealed that Daidzina presents better interaction between the ligands under analysis. The interaction of Daidzina with the receiver PDB-6IUI is mediated mainly by the amino acid residues MET112, THR298, LEU294, THR81, CYS35, interactions that could be targets therapeutics for inhibiting the formation of thromboxane A2.
Nitric oxide, prostacyclins and heparin inhibit platelet function and thrombus formation through the platelet receptor IP (G6b- B) PDB- 6R0X.
In the study, the results of the interaction of each ligand, estradiol and isoflavone molecules with the platelet receptor G glycoprotein IP (G6b-B), showed that O Binder Estradiol has greater selectivity and affinity with the receptor than isoflavones, presenting good interaction, which is mediated mainly through the amino acid residues GLY77, VAL78, THR73, this interaction reduces the risk of thrombus formation, shown here for Estradiol isoflavone molecules with the IP platelet receptor (CLEC2) PDB-4YK5, Estradiol in comparison with isoflavones showed greater interaction with the receiver. The interaction of Estradiol with the receptor PDB-4YK5 is mediated mainly due to the amino acid residue ARG122, which also interacts with isoflavone molecules, making the ARG122 residue for this receptor, an option for drug action with the aim of minimizing the risk of thrombosis for these ligands.
The results of the interaction of each ligand with the platelet receptor tyrosine kinase P2Y1 and ATP PDB-4XZ2, Estradiol in comparison with isoflavones showed a greater interaction, which it is mainly mediated by the amino acid residue CYS98, ALA96. These receptors, when activated, cause changes in the shape and aggregation of platelets, production of thromboxane A2, and adhesion of fibrinogen, inducing thrombus formation. For Estradiol, the cysteine amino acid residue (CYS97) also serves as an interaction for the P2Y12/ADP PDB-4PXZ receptor. Pharmacological action on this residue could minimize the risk of thrombosis when using Estradiol for menopausal hormone replacement.
To the platelet receptor tyrosine kinase Syk/CLEC2 PDB-3IET, shows that among the ligands in the study, the isoflavone Daidzine has the greatest selectivity and the highest affinity by the platelet tyrosine kinase receptor Syk/CLEC2, which is mainly mediated by the amino acid residue ARG98, SER91. For this receptor, ARG52 residues are present in all interactions with the other ligands under analysis. These receptors, when activated, cause changes in the shape of platelets in the platelet aggregation process for thrombus formation, generation of thromboxane A2 and fibrinogen adhesion.
This work highlights the importance of using anchoring, extensively cited in the literature, as an essential tool in the design of drugs based on molecular structure, as well as used to optimize known drugs and to identify new ligands by predicting their binding mode and affinity at interaction sites. Furthermore, these methods are fast enough to allow virtual screening of ligand libraries leading to in silico studies based on experimental studies.
Concluding that the ligands studied, estradiol and isoflavones (Daidzein, daidzin, genistein, genistin, glycitein, glicitin), had good interaction with platelet receptors.
Estradiol showed good interaction and good affinity with the 2GI7 (GPIV) receptor by hydrogen bonding through ARG67, and by van der Waals bonds through the amino acids TRP76, SER69, ARG38, GLU40, PRO79, CYS68, LEU42, LEU39. The platelet activation response is an increase in fibrin, stabilizing the clot. In contrast to this interaction, Estradiol also showed good interaction and good affinity with the receptor with 6R0X (IP G6b-B) by hydrogen bonding through GLY77, VAL78, THR73, and by van der Waals bonds through the HIS75 amino acids, LEU74, which responds to the inhibition of platelet activation. Therefore, based on the results of the interactions under study, the Estradiol ligand has action on platelet activation receptors and platelet inhibition receptors.
And among isoflavones Daidzina interacted with a greater number of platelet receptors. And, like Estradiol, it interacted better with the 2GI7 receptor (GPIV) by hydrogen bonding through the amino acids ASP49, GLN50, TYR47, ALA57, and by van der Waals bonds through the amino acids GLN48, AGN46, LEU53, ILE55, PRO56, LEU62.
In contrast to this interaction, like Estradiol, it also showed good interaction and good affinity with the 6R0X receptor (IP G6b-B) by hydrogen bonding through GLY77, LEU74, THR73, SER72, TYR70, ASN51, and by van der Waals bonds through the amino acids HIS75, SER76.
Among isoflavones, Genistein showed less interaction and lower affinity for a smaller number of platelet receptors under study.
New drugs can be developed by intercepting interactions, and molecular modeling studies can help improve the choice of drug used in replacement therapy during menopause, minimizing cardiovascular risks and improving women's quality of life.
Conclusion
Estradiol and Daidzine has better interaction with this 2G17 receptor, resulting in greater platelet activation and greater capacity for thrombus consolidation in relation to the other molecules under study. And of the 14 platelet receptors under study and identified in the PDB, only 04 (1P8V; 6IIU; 4YK5; 3IET) presented crystallographic structure of the ligands. Estradiol has good selectivity and affinity with the 6R0X receptor, therefore, providing some protection against thrombus formation upon interaction with the receptor.
Funding
The authors do not have any conflict of interests. This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
References
- Calf IMP. Translational medicine and contributions to public health. J Hum Growth Dev. 2017;27(1):6-9.
- Wooller SK, Benstead-Hume G, Chen X, Ali Y, Pearl FMG. Bioinformatics in translation drugs discovery. Biosci Rep. 2017;37(4).
[Crossref] [Google Scholar] [PubMed]
- Lupatini EO, Barreto Ivan JOM, Zimmermann R, Silva EM. Medicines and translational research: steps, actors, and health policies in the Brazilian context. Saúde Debate. 2019;43:1-10.
- Liang X, Zhu W, Lv Z, Zou Q. Molecular Computing and Bioinformatics. Molecules. 2019;24(13):2358.
[Crossref] [Google Scholar] [PubMed]
- Watson SP, Harrison P, Halford GM. Platelets: the next decade. Platelets. 2020;31(1):1-2.
[Crossref] [Google Scholar] [PubMed]
- Singh S, Baker QB, Singh DB. Molecular docking and molecular dynamics simulation. 2022;18:291-304.
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, et al. Automated anchoring using a Lamarckian genetics algorithm and an empirical binding free energy function. J Comput Chem. 1998;19(14):1639-1662.
- Jamal QMS, Khan MI, Alharbi AH, Ahmad V, Yadav BS. Identification of natural compounds of the apple as inhibitors against cholinesterase for the treatment of Alzheimer's disease: an in silico molecular anchorage simulation and ADMET study. Nutrients. 2023;15(7):1579.
[Crossref] [Google Scholar] [PubMed]
- Breton R, Housset D, Mazza C, Fontecilla-Camps JC. The structure of a complex of human 17beta-hydroxysteroid dehydrogenase with estradiol and NADP+ identifies two main targets for the design of inhibitors. Structure. 1996;4(8):905-915.
[Crossref] [Google Scholar] [PubMed]
Citation: Soares VM, Fonseca AM, Fonseca O, Seabra M, Ramos DMAM, Nascimento LT, et al. (2025) Estrogen Agonist Induced Effects on Platelet Activation Properties and Platelet Signaling Pathways by Structural Class. Adv Pharmacoepidemiol Drug Saf. 14:391.
Copyright: © 2025 Soares VM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.