Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- CiteFactor
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research - (2022) Volume 13, Issue 3
Efficacy and Durability of Paenibacillus sp. strain B2 in Co-Inoculation with Arthrobacter sp. SSM-004 and Microbacterium sp. SSM-001 for Growth Promotion and Resistance Induction in Wheat against Mycosphaerella graminicola and Drought Stress
Erika Samain1,2, Cédric Ernenwein2, Thierry Aussenac3 and Sameh Selim1*2SDP, 1 rue Quesnay, 02000 Laon Cedex, France
3Institut Polytechnique UniLaSalle, Université d’Artois, ULR 7519, 19 Rue Pierre Waguet, BP 30313, F-60026 Beauvais Cedex, France
Received: 16-Mar-2022, Manuscript No. JPPM-22-15796; Editor assigned: 18-Mar-2022, Pre QC No. JPPM-22-15796 (PQ); Reviewed: 01-Apr-2022, QC No. JPPM-22-15796; Revised: 08-Apr-2022, Manuscript No. JPPM-22-15796 (R); Published: 18-Apr-2022
Abstract
Plant Growth-Promoting Rhizobacteria (PGPR) are able to promote plant growth and/or induce local and systemic resistance against biotic and abiotic stresses, but the stability and durability of their efficiency still need more investigation. The present work aims to identify a compatible-PGPR-mixture effective to stimulate wheat growth, resistance against Mycosphaerella graminicola, the causal agent of Septoria tritici leaf Blotch (STB), and tolerance to drought stress. The interactions between twenty-six PGPR and four wheat cultivars with different resistance levels to STB, in individual and co-inoculations, were tested. The results demonstrated higher external and internal root colonisation potential of Paenibacillus sp. strain B2 (PB2) in a mixture, referred hereafter as Mix-3, with strains Arthrobacter sp. SSM-004 and Microbacterium sp. SSM-001, and without an impact of wheat genotype and growth stage, as observed in its individual inoculations. Only with Mix-3 was wheat growth promotion observed. Interestingly, PB2 and Mix-3 eliminated the negative impact of drought stress on the Foliar Dry Biomass (FDB) and Root Dry Biomass (RDB), and only Mix-3 impacted root length. Moreover, Mix-3 induced pathogen strain and growth stage-dependent resistance, and conferred more than 73.5% protection against STB compared to 59.8% by PB2 in a single inoculation. Gene expression results showed the activation of basal defences, reactive oxygen species, phenylpropanoid and phytoalexins, salicylic acid and jasmonic acid pathways in the resistance induced by Mix-3. The PR1, chitinase, glucanase and flavonoid genes are strongly recommended as protection gene markers for wheat resistance to STB. To conclude, PB2 induced durable wheat resistance against M. graminicola and wheat tolerance against drought stress. Only in a mixture of three-compatible-PGPR (Mix-3) was plant growth promotion observed and the tolerance induced to drought stress was more effective. However, it seems that resistance induced against STB is PB2-dependent.
Keywords
Mycosphaerellagraminicola; Drought stress; Induced systemic resistance; Plant growth-promoting rhizobacteria; Paenibacillus sp. strain B2; Arthrobacter sp. SSM-004; Microbacterium sp. SSM-001
Introduction
Plants interact with a highly diverse range of microorganisms, establishing symbiotic interactions that may play an important role in plant protection against environmental stresses by enhancing plant defences [1-4]. Beneficial microorganisms such as Plant Growth-Promoting Rhizobacteria (PGPR), present in the plant rhizosphere, are able to colonise roots and thereby improve plant growth and crop yield via N2 fixation, phosphate solubilisation and phytohormone production, by limiting plant pathogen impacts through the production of antimicrobial compounds, and by stimulating mechanisms of Induced Systemic Resistance (ISR) in the presence of biotic and abiotic stresses [2,5]. The major impact of ISR is its quantitative and non-specific resistance, providing protection against a large spectrum of stresses; this may be useful in stress management in agriculture caused by climate change, pathogen resistance emergence, or the limited use and efficiency of pesticides [6]. Systemic resistance can be triggered after pathogen infection, treatment using chemicals, or root colonisation by PGPR and is characterised by the elicitation of plant defence mechanisms, mediated by the Jasmonic Acid (JA) and Ethylene (ET) pathways, involving PR proteins, phytoalexin synthesis, and the formation of physical barrier [7]. Recently, studies have also demonstrated a role of PGPR against drought stress by enhancing water and nutrient uptake, reducing oxidative damage, and improving plant growth [8], using several mechanisms such as Induced Systemic Tolerance (IST), similar to ISR and defined as the physical and chemical changes enhanced by plant-microbe interactions that enhance plant tolerance to abiotic stresses [9-11].
However, several studies have revealed the inconsistent efficacy of individual PGPR inoculation and demonstrated enhanced protection with a consortium of PGPR, providing synergistic activities against a wide range of pathogens [12]. Mixtures of PGPR are a solution to increase growth by direct and indirect mechanisms, and through the control of biotic and abiotic stresses.
The PGPR Paenibacillus sp. strain B2 produces the cyclic lipo- polypeptide antibiotic paenimyxin [13], a well-known as systemic resistance inducer against fusarium root rot in Medicago truncatula and STB [2,4,7]. In single inoculation, PB2 has no promoting impact on plant growth and has demonstrated effective but cultivar-dependent protection against Mycosphaerella graminicola, the causal agent of Septoria tritici leaf Blotch (STB) [2].
In the present study, our objective was to determine a PGPR mixture able to enhance wheat growth and resistance against drought and STB stresses. Pathogen strains, plant genotypes, and growth stages were taken into consideration in the selection of this PGPR mixture. The expression of gene markers of the most important plant defence pathways was studied.
Materials and Methods
Microorganisms and inoculum preparation
Four M. graminicola strains were used in this study, i.e., strain IPO323 (provided by Dr. F. Suffert, INRA-AgroParisTech (Laboratory of Phytopathology), strain 1193, characterised as a moderately resistant strain to DMI fungicides with three SNP mutations (M-281-V, A-379-G, I381-V) (Selim, 2017, NCBI Gene Bank database accession number KX356102), and strains TO256 and ST38 from the authors’ laboratory. Twenty-six PGPR were selected, including Paenibacillus sp. strain B2 [14], kindly provided by Dr. D. van Tuinen (INRA Dijon, France) and Paraburkholderia phytofirmans PsJN (provided by Dr. E. Ait Barka, URCA, Reims, France). Eleven were from DSMZ Germany (DSM1-11), and 13 were from the authors’ laboratory (EDS, SSM-001-012) (Table 1).
| Abbreviation | Title | Gene bank accession n° |
|---|---|---|
| PB2 | Paenibacillus sp. strain B2 | AJ011687.1 |
| PSjN | Paraburkholderia phytofirmans PsJN | AY497470 |
| Cp | Curtobacterium plantarum strain EDS | KX458115 |
| DSM-1 | Azotobacter chroococcum DSM 2286 | |
| DSM-2 | Azospirillum brasilense DSM 1690 | |
| DSM-3 | Arthrobacter oxydans DSM 20119 | |
| DSM-4 | Enterobacter cloacae subsp. dissolvens DSM 16657 | |
| DSM-5 | Bacillus megaterium DSM 32 | |
| DSM-6 | Paenibacillus polymyxa DSM 36 | |
| DSM-7 | Bacillus amyloliquefaciens subsp. amyloliquefaciens DSM 7 | |
| DSM-8 | Bacillus pumilus DSM 27 | |
| DSM-9 | Pseudomonas chlororaphis subsp. aureofaciens DSM 6698 | |
| DSM-10 | Rhizobium radiobacter DSM 30147 | |
| DSM-11 | Streptomyces cellulosae DSM 40362 | |
| SSM-001 | Microbacterium sp. strain SSM1 | MF574166 |
| SSM-002 | Curtobacterium plantarum strain SSM2 | MF574170 |
| SSM-003 | Agrobacterium sp. strain SSM3 | MF574177 |
| SSM-004 | Arthrobacter sp. strain SSM4 | MF574214 |
| SSM-005 | Raoultella sp. strain SSM5 | MF574215 |
| SSM-006 | Xanthomonas sp. strain SSM6 | MF574216 |
| SSM-007 | Pseudomonas sp. strain SSM7 | MF574217 |
| SSM-008 | Rahnella aquatilis strain SSM8 | MF574218 |
| SSM-009 | Rahnella aquatilis strain SSM9 | |
| SSM-010 | Rahnella aquatilis strain SSM10 | |
| SSM-011 | Pseudomonas fluorescens SSM13 | MF574219 |
| SSM-012 | Pseudomonas sp. SSM14 | MF574220 |
Table 1: List of the plant-growth-promoting rhizobacteria tested.
PGPR inocula were prepared as described by Selim et al [13]. Briefly, to carry out the final bacterial inocula, bacterial cells were pelleted from Luria-Bertani (LB) liquid cultures at optical density OD 0.5, by centrifugation at 2655×g for 10 minutes and at 4°C, washed three times and then suspended in a sterile solution of 10 mM MgSO4. M. graminicola inocula were prepared as described by Selim et al. [15]. Briefly, sporidia stored at -80°C were transferred to potato dextrose agar medium. After 7 days of incubation at 18°C with a 12 hour photoperiod, mycelia and spores were scraped from the surface and transferred into a liquid yeast-sucrose medium for 5 days at 18°C with permanent light (100 µmol of photon m-2 s-1) and shaking (150 rpm). Spores were collected by centrifugation at 2655×g for 10 min at 15°C, washed three times with a sterile solution of 10 mM MgSO4 and then suspended in 10 mM MgSO4, containing 0.05% Tween 20 as a surfactant. The final concentration was adjusted to 106 spores.mL-1.
Plant material and growth conditions
Four wheat cultivars, i.e., Alixan, Altigo, Cellule, and Hyfi, with different levels of resistance against STB (4, 5.5, 6.5, and 7, respectively, on a scale from 1 (fully susceptible) to 9 (fully resistant)) were used in this study (Table 2). Grains were disinfected according to Samain et al., with a few modifications, as follows: incubation in a solution of oxytetracycline, streptomycin, penicillin, and ampicillin antibiotics (100 mg.L-1 each) overnight to obtain broadspectrum activity against bacterial strains, then suspended in 10% calcium hypochlorite solution for 10 minutes and washed three times in autoclaved Milli-Q water after each disinfection step. The sterilised grains were pre-germinated on 0.5% water-agar medium and incubated in darkness at 4°C for 24 h, 20°C for 48 h, and 4°C for 24 h. Germinated grains were transferred into an inoculum of individual PGPR or in a PGPR mixture with an equal quantity of each. Inocula for single or co-inoculation were adjusted to a final concentration of 106 CFU (Colony Forming Units).mL-1 of 10 mM MgSO4. One millilitre per grain was used for one hour with light shaking. For the non-inoculated control, grains were immersed in 10 mM MgSO4. After inoculation, grains were transferred into 250- ml pots containing a sterilised soil mixture of silt-loam soil and sand (1:1, v/v). Pots were incubated in a phytotron at 18°C (+/- 2°C), 40% humidity, for a 16-hour photoperiod with 185 μmol m-2 s-1 photon flux density supplied by high-output white fluorescent tubes (Philips Master Cool White 80 W/865, Lamotte Beuvron, France). Plants were watered three times a week with 50 ml of distilled water per pot.
| Cultivar | Producer | Year | Susceptibility rating* |
|---|---|---|---|
| Alixan | LG | 2005 | 4 |
| Altigo | LG | 2007 | 5.5 |
| Cellule | Florimond desprez | 2012 | 6.5 |
| Hyfi** | Saaten Union | 2013 | 7 |
Note: *The susceptibility rating is on a scale of 1 to 9, where 1 represents ‘highly susceptible’ and 9 represent ‘highly resistant’. **Hybrid cultivars.
Table 2: Wheat cultivars used to study the impact of wheat genotypes on the resistance induced by mix-3.
Screening PGPR for root colonisation
In a single inoculation, the external and internal root colonisation by the 26 studied PGPR were analysed by counting CFU as described by Samain et al. [2] at 7 days after inoculation (dai) and sowing (das) on water agar plates, using the Alixan, Altigo, Cellule, and Hyfi wheat cultivars.
Screening PGPR for the induced resistance against STB
Wheat seed sterilisation, pre-germination, inoculation with PGPR, and phytotron conditions were carried out as mentioned above. Protection against STB and plant growth promoting effects was used to evaluate the 26 studied PGPR in a single inoculation. The four cultivars, i.e., Alixan, Altigo, Cellule, and Hyfi, were inoculated with M. graminicola strain 1193 (106 spores/leaf), at the three-leaf growth stage. The infection level was determined at 17 dai using qPCR as described by Selim et al. [15].
Wheat root colonisation with PGPR mixture
The external and internal root colonisation by the selected PGPR from the screening results (Paenibacillus sp. strain B2 (PB2), Microbacterium sp. strain SSM1 (SSM001) and Arthrobacter sp. strain SSM4 (SSM004)), composing Mix-3, were quantified at 21 das (three-leaf growth stage (3-L GS)) on the four cultivars Alixan, Altiog, Cellule, and Hyfi. External and internal colonisation by these PGPR were also determined at flag leaf growth stages (F-L GS) on Alixan and Cellule, using 16S rDNA-specific primers, designed by Dr. Sameh Selim, by real time quantitative qPCR, as described by Samain et al. (Table 3). Briefly, DNA was extracted from plant roots using a DNeasy 96 Plant kit (Qiagen, USA), according to the manufacturer’s protocol. DNA quantity and quality were confirmed on a Nanodrop apparatus (Thermo Fisher Scientific, Waltham, MA, USA). SYBR Green qPCR assays were carried out in a reaction mixture of 25 µL that contained the following: 12.5 µL Universal Quantifast SYBR Green PCR master mix (Qiagen, USA), 0.3 µM of each primer, 50 ng of DNA, and water up to a volume of 25 µL. The conditions of quantitative PCR were as follows: 5 min at 95°C, followed by 40 cycles of 10 s at 95°C and 30 s at 60°C. All quantitative PCR was carried out using Step One Plus (Thermo Fisher Scientific®).
| Gene name | Gene bank accession N° | For/Rev primers (5’-3’)** | Tm* (°C) | Amplicon length (bp) | MT* (°C) | PCR efficiency (%) |
|---|---|---|---|---|---|---|
| Housekeeping genes | ||||||
| Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) | AF251217 | AAGGGCATTTTGGGTTACGTT | 58.4 | 63 | 79.1 | 103.98 |
| CCTGTTGTCACCCTGGAAGTC | 58.1 | |||||
| β-tubulin (Β-TUB) | U76897 | CTGCCTCCAAGGTTTCCAAGTA | 59.2 | 63 | 81 | 92.7 |
| GTGTCCATCCCGGAACCA | 58.7 | |||||
| Cell wall proteins and basal defense | ||||||
| Pathogenesis-Related Protein (PR1) | HQ848391 | CATGCACCTTCGTATGCCTAACT | 58.9 | 52 | 79.1 | 90.25 |
| TGGCTAATTACGGCATTCCTTT | 59.4 | |||||
| Chitinase (CHIT) | AB029935 | GGGTGGACCTGCTGAACAAT | 58.4 | 75 | 84.6 | 92.35 |
| AGAACCATATCGCCGTCTTGA | 58.3 | |||||
| β-1,3-Glucanase (GLUC) | DQ090946 | TCCTGGGTTCAGAACAATGTCC | 59.8 | 50 | 78.2 | 105.35 |
| TTGATGTTGACAGCCGGGTAGT | 60.4 | |||||
| Thaumatin-Like Protein (TLP) | CD86039 | AGGTAATTTTTTTATTGCCCTGTACTG | 58.9 | 89 | 77.7 | 90.25 |
| TTACAGCCGCCGTACTACATGT | 60.3 | |||||
| JA signaling pathway | ||||||
| Lipase (LIP) | TaBs117A2 | Cacaaaatatcgacccaccac | 60 | 149 | 86.3 | 100.92 |
| actgggtattcgtctgtcagc | 59 | |||||
| Lipoxygenase (LOX) | U32428 | GGGCACCAAGGAGTACAAGGA | 59.9 | 66 | 82.2 | 99.66 |
| GCTCGTGATGGTGTGGATGA | 59.1 | |||||
| Allene Oxide Synthase (AOS) | AY196004 | AGGCCGGAGAGAAGTTCCAC | 59.3 | 119 | 88 | 93.65 |
| CCGACTTGGTCAGCTCCATC | 59.2 | |||||
| Phenylpropanoid and phytoalexin pathway | ||||||
| Phenylalanine Ammonia-Lyase (PAL) | AY005474 | GTCGATTGAGCGTGAGATCAAC | 58 | 59 | 80.5 | 101.35 |
| CACGGGAGACGTCGATGAG | 59 | |||||
| Chalcone Synthases (CHS) | AY286097 | GCGCCTGCGTACTCTTCATC | 60 | 51 | 80.8 | 109.67 |
| CCTCGGCGGAGCGTTT | 59 | |||||
| Flavonoid 7-O-methyltransferase-like (FLAV) | CA682712 | GACAACAAGGAGGCTGTGTATGG | 62.4 | 117 | 80.6 | 93.43 |
| GGTGTAATGCAGTTGAATCAAGGA | 59.3 | |||||
| Reactive Oxygen Species (ROS) | ||||||
| Peroxidase (POX) | X85228 | TGCTTTGTCCAAGGCTGTGA | 59 | 61 | 79.8 | 108.68 |
| GACCCGCGTTTTGTTCCA | 59.1 | |||||
| Oxalate Oxidase (OXO) | AJ556991 | GCCAGAACCCCGGTATCG | 60 | 55 | 80.8 | 97.23 |
| GGTGGGTTGGAGCTGAAGAG | 58 | |||||
| Glutathione-S-Transferase (GST) | AF397085 | CGCTCTGAGCCCCATTCTC | 59.5 | 55 | 79.1 | 106.28 |
| GGCTCCCCCAAGCATAGG | 59 | |||||
| Germin-Like-Protein (GLP) | Y09916 | AGGTGAGCTCCTTGTTGGAATC | 60.3 | 121 | 85.6 | 91.99 |
| GTTGAACTGGAAGTGCATGAGG | 60.3 | |||||
| Glutathione Peroxidase (GPX) | KM817777 | GTTCAGTTTGCCTGCACTCG | 58.1 | 141 | 83.3 | 94.17 |
| GTTCCACTTGATGCTGTCGC | 57.9 | |||||
| Catalase (CAT) | X94352 | TTCAAGCAGGCTGGTGAGAG | 59.8 | 106 | 84.5 | 103.09 |
| TTTCATGGGTGACACGAGCA | 59.7 | |||||
| Superoxide Dismutase (SOD) | EF392662 | TCAGGACCCTCTTGTGACCA | 57.6 | 100 | 81.7 | 102.65 |
| CGGCCTCACGTTCTTGTACT | 56.6 | |||||
| Defense and cell rescue | ||||||
| related protein Kinase (rpK) | KR611569 | TTTTGTTGGGGATCCTGCGT | 61.2 | 128 | 81.5 | 99.66 |
| GCTCAGGCTCCTCGTATTGG | 58.5 | |||||
| WRKY1 transcription factor (WRKY) | EU665424 | TGGCGCAAGTATGGTCAGAA | 58.9 | 77 | 79.3 | 101.35 |
| CAGCCCTGGTGGGTACATTT | 58.4 | |||||
| MAP kinase (WCK1) | AF079318 | AGTTCGAGATCACGGCCAAGT | 59.8 | 131 | 87.6 | 108.19 |
| GAAGGCGTTGGCGATCTTC | 58.8 | |||||
| 16S ribosomal DNA (16S rDNA) | ||||||
| Paenibacillus sp. strain B2 | AJ011687 | TCGTAAAGCTCTGTTGCCAGG | 59 | 51 | 75.45 | 109.67 |
| CTTGAGCAGTTACTCTACAAGACGTTC | 58 | |||||
| Microbacterium spp. | AACACCATCAACACCCACGA | 57.3 | 222 | 88.4 | 93.25 | |
| CCTCGGTGTTGCCGAGCTT | 53 | |||||
| Arthrobacter spp. | GATCTGCGGTGGGTACGG | 56.5 | 380 | 86.91 | 98.43 | |
| CGGTTCATGTCAAGCCTT | 53.7 | |||||
| Note: * Tm, primer’s annealing temperature; MT, amplicon’s specific melting temperature. ** Primers’ reference, Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH), β-tubulin (Β-TUB); Pathogenesis-Related Protein (PR1), Chitinase (CHIT), β-1,3-Glucanase (GLU), Lipoxygenase (LOX), Allene Oxide Synthase (AOS), Phenylalanine Ammonia-Lyase (PAL), Chalcone Synthases (CHS), Peroxidase (POX), Oxalate Oxidase (OXO) and Glutathione-S-Transferase (GST) [16]; Thaumatin-Like Protein (TLP), Flavonoid 7-O-methyltransferase-like (FLAV), and Germin-Like-Protein (GLP); Lipase (LIP); MAP kinase (WCK1); Glutathione Peroxidase (GPX), Catalase (CAT), Superoxide Dismutase (SOD), related protein Kinase (rpK), WRKY1 transcription factor (WRKY), and 16S rDNA [7]. Primer pairs were designed using the Primer Express® program and tested for secondary structure using the AmplifX® program. All used primers did not show any form of dimerization. | ||||||
Table 3: Oligonucleotide primer sequences of wheat-defense genes and the 16S rDNA for Paenibacillus sp. strain B2, Microbacterium sp. SSM-001, and Arthrobacter sp. SSM-004.
The standard curve, obtained by plotting known amounts of each bacterium DNA against Ct values, was used to determine the amplification efficiency (Table 3) (Supplementary Figures 1-3). The resulting regression equations were used to calculate the amounts of PB2, SSM001 and SSM004 DNA in root DNA samples.
Mix-3 growth promotion effect on wheat
The impact of PGPR on wheat biomass was evaluated 6 weeks after the inoculation of germinated grains by measuring root and foliar dry biomasses (RDB and FDB, respectively) of the Alixan, Altigo, Cellule, and Hyfi cultivars.
Mix-3 resistance induced in wheat against drought stress
The induction of resistance against drought stress in wheat by PB2 and Mix-3 was studied in a phytotron, using Alixan, Altigo, Cellule, and Hyfi wheat cultivars, as a response to the inoculation of the pre-germinated grains with PB2 and Mix-3 at sowing, as mentioned above. Plants were watered with 50 mL/pot twice a week, for 2 weeks, followed by 19 days without watering for the drought stress modalities. To study the plant growth recovery capacity, the drought stressed period was followed with 2 weeks of normal watering with 50 ml/pot twice a week. Foliar and root dry biomass, as well as root length, were measured at the end of the recovery period.
Mix-3 resistance induced in wheat against M. graminicola
The four cultivars Alixan, Altigo, Cellule, and Hyfi were used to study the stability of the protective efficiency of PGPR Mix-3 compared to individual inoculation with PB2. Plants were inoculated with M. graminicola strain 1193 or the green fluorescent protein gene (GFP)-recombinant strain IPO323 at 3-L GS by spraying 2 ml of M. graminicola inoculum (2 × 106 spores/mL) over the whole plant. Controls were sprayed with a solution of 10 mM MgSO4 containing 0.1% Tween 20 as a surfactant. Seventeen days after infection with M. graminicola, leaves were collected and lyophilised to evaluate the protection efficiency as a response to PB2 or Mix-3 using qPCR. The DNA extraction and quantification of M. graminicola using qPCR was performed as described by Selim et al. [14]. Briefly, the DNA was extracted from plant leaves using a DNeasy 96 Plant kit (Qiagen, USA), according to the manufacturer’s protocol. DNA quantity and quality were confirmed by Nanodrop (Thermo Fisher Scientific, Waltham, MA, USA). To quantify infection levels of M. graminicola, primers and a TaqMan minor groove binder probe (For: GCCTTCCTACCCCACCATGT; Rev: CCTGAATCGCGCATCGTTA; Probe: FAM-TTACGCCAAGACATTC-MGB) were used to target a 63-bp fragment of the M. graminicola β-tubulin specific gene (Gene Bank accession no. AY547264) [16,17]. A TaqMan assay was carried out in a 25 µL reaction mixture that contained 12.5 µL of Universal TaqMan PCR Master Mix (Life Technologies SAS, Villebon sur Yvette, France), 0.3 µM of each primer, 0.2 µM of probe, 200 ng of DNA, and water to a volume of 25 µL. The conditions for qPCR determination were as follows: 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. All qPCR experiments were carried out using a Step One Plus Real-Time PCR System (Thermo Fisher Scientific®). qPCR analysis of the M. graminicola β-tubulin gene was calibrated from 102 to 107 copies by serial dilution of the appropriate cloned target sequence, as previously described [18,19].
Leaf colonisation and pycnidia formation were observed using an Axioscope A1 binocular magnifier and epifluorescence microscope (ZEISS, Germany), at 3 and 21 dai, respectively.
Stability and durability of Mix-3 resistance induced over wheat genotypes, growth stages, and pathogen strains
Susceptible and resistant wheat cultivars, i.e. Alixan and Cellule, respectively, were used to evaluate the effect of pathogen strains and growth stage on the efficiency of the resistance induced by Mix-3. They were inoculated with the four M. graminicola strains at 3-L, Tillering (Ti), and F-L GS. The inocula of each strain were prepared and applied as mentioned above. Five repetitions were carried out for each condition. Three control modalities were used as non-infected with M. graminicola and non-inoculated with Mix-3 (C-), inoculated with Mix-3 without pathogen infection (Mix- 3), and non-inoculated with Mix-3 infected with pathogen (MG). Modalities inoculated with Mix-3 and infected with M. graminicola (Mix-3/MG) were compared to control modalities. Seventeen days after infection with M. graminicola, leaves were collected and lyophilised to evaluate the protection efficiency as a response to Mix-3 using qPCR as mentioned above.
RNA extraction and relative gene expression quantification by real-time PCR
At the 3-L GS, aerial parts of Alixan and Cellule plants were collected at the Time of Infection (T0) with M. graminicola, at 6, 12, 24, and 48 hours after infection (hai), and at 3, 5, 9, and 11 dai to study the evolution of defence gene expression. Samples were stored directly in liquid nitrogen. RNA extraction and cDNA synthesis were carried out using an RNeasy® Mini Kit and QuantiTect® Reverse Transcription Kit (Qiagen, USA), respectively, following the manufacturer’s protocol. The gene expression of 20 wheat defence genes was studied using specific primers (Table 3). qPCR conditions were as described by Samain et al. Briefly, the Quantifast® SYBR® Green PCR Kit (Qiagen, USA) and the StepOnePlus Real-Time PCR System (Thermo Fisher Scientific®) were used. Amplification conditions consisted of a denaturation cycle (95°C for 5 minutes) and amplification and quantification cycles (95°C for 10 seconds, 60°C for 30 seconds) repeated 40 times. One final step from 60°C to 95°C with an increase of 0.2°C s-1 was added to obtain a specific denaturation curve for each studied gene (Table 3). The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-tubulin (β-TUB) housekeeping genes were used to normalise results and to determine the expression ratio for each cDNA, as described by Ors et al [16]. Briefly, expression ratios for each cDNA were calculated for each time point, relative to control at the same time using the 2-ΔΔCt method described by Livak and Schmittgen [45], where ΔΔCt=[Ct Target (Sample) – Ct Reference (Sample)]-[Ct Target (Control) – Ct Reference (Control)] and Ct Reference=geometric mean (Ct GAPDH:Ct β-TUB). Similar amplification efficiencies ranging between 90% and 110% were checked for all the tested primers (Table 3) and expression ratio values of 2 were considered as a minimum to be significantly different from the control.
Statistical analysis
Each experiment was repeated at least three times and contained at least five replicates. For all experiments, significant differences were evaluated using the Tukey test at p ≤ 0.05 with the XLSTAT® statistics program (version 2014, Addinsoft, Paris, France).
Results
Screening PGPR
Twenty-six PGPR, in individual inoculation of germinated wheat grains, as mentioned above, were screened for their potential of external and internal root colonisation, inducing protection against M. graminicola and increasing plant biomass. Four wheat cultivars with different levels of resistance against STB were used.
Screening for wheat root colonisation
The results in Figure 1 and Supplementary Table 1 shows that PB2, PSjN, Cp, DSM-2, DSM-3, DSM-4, DSM-10, SSM-001, SSM-002, SSM-003, SSM-004, SSM-005, SSM-006, SSM-010, and SSM-012 had excellent potential (more than 106 CFU/g of root) for external root colonisation. Internal colonisation depended on wheat genotype for PB2, PSjN, DSM-1, DSM-4, DSM-5, DSM-7, DSM- 8, DSM-10, DSM-11, SSM-002, SSM-004, SSM-007, and SSM-012 (Figure 1) (Supplementary Table 1). For DSM-6, DSM-9, SSM-001, SSM-006, and SSM-009, PGPR were not endophytic in all tested cultivars.
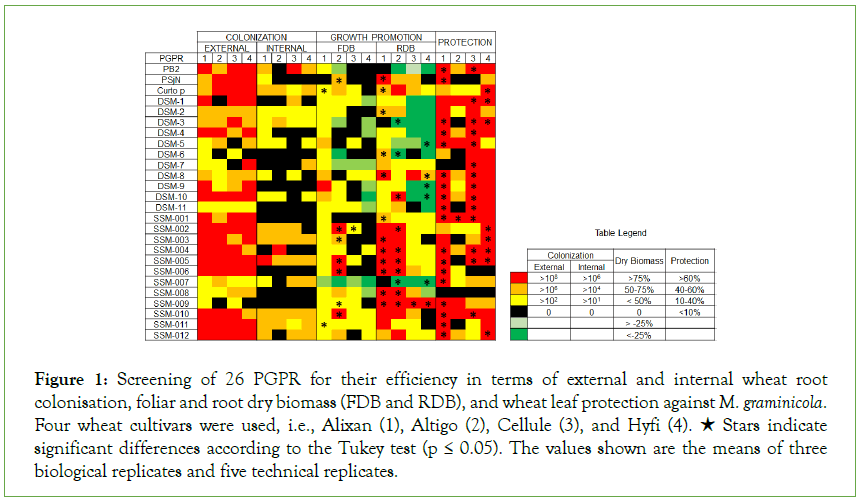
Figure 1: Screening of 26 PGPR for their efficiency in terms of external and internal wheat root colonisation, foliar and root dry biomass (FDB and RDB), and wheat leaf protection against M. graminicola. Four wheat cultivars were used, i.e., Alixan (1), Altigo (2), Cellule (3), and Hyfi (4). ★ Stars indicate significant differences according to the Tukey test (p ≤ 0.05). The values shown are the means of three biological replicates and five technical replicates.
Screening for wheat growth promotion
For wheat growth promotion, some of the screened PGPR seemed to have a negative impact on FDB and RDB. Indeed, the PGPR DSM-3, DSM-5, DSM-6, DSM-9, DSM-10, and SSM-007 led to a significant decrease in general wheat growth. On the other hand, PSjN, Cp (EDS), SSM-002, SSM-003, SSM-005, SSM-006, and SSM-009 showed a significant increase in both FDB and RDB. Other PGPR such as DSM-2, DSM-8, SSM-001, SSM-004, and SSM-008 promoted only root growth and SSM-010 and SSM-011 increased only FDB for some and not all tested cultivars (Figure 1 and Supplementary Table 1).
Screening for wheat induced resistance against STB
High protection levels were observed in almost tested PGPR, but with important variations between wheat cultivars (Figure 1) (Supplementary Table 1). For example, the PGPR PSjN did not show any protective effect on Altigo and Cellule but provided 70% protection efficiency on Alixan. Likewise, DSM-3, DSM-6, DSM-7, DSM-11, SSM-003, SSM-005, and SSM-011 did not show any protection efficiency against M. graminicola for Altigo, as with DSM-7 for Alixan, and SSM-006 for Cellule and Hyfi. In only one case, no protection induction was observed for the four tested cultivars when roots were inoculated with the PGPR SSM-008. However, PB2, DSM-1, DSM-4, DSM-8, DSM-10, SSM-001, SSM-004, SSM-009, and SSM-010 provided strong and stable protection efficiencies, more than 40%, for the four tested cultivars.
Based on these results, PGPR with negative effects on wheat biomass and/or without or with a weak protective effect against STB were excluded. However, PB2, DSM-8, SSM-001, SSM-004, SSM-009, and SSM-010 were selected for studying their growth promoting and protection efficiency in co-inoculation. After the analysis of biomass stimulation and protection inducing potential of all the possible mixtures between these selected PGPR (data not shown), a mixture of three PGPR, i.e., PB2, SSM-001, and SSM- 004, was chosen and referred to as Mix-3.
Impact of root colonisation by Mix-3 on the PB2 colonisation level
The results in Table 4 shows an increase in internal root colonisation with PB2, between 3.6 to 18.6 times, in Alixan, Cellule, and Hyfi cultivars inoculated with Mix-3, compared to the colonisation level after single inoculation with PB2. However, no modification in the PB2 colonisation level was observed for Altigo. Similarly, external colonisation with PB2 was increased by 3.4, 6.9, and 1.5 times in Altigo, Alixan, and Hyfi, respectively, and was stable in Cellule, compared to single inoculation (Table 4).
| Colonisation | ||
|---|---|---|
| Wheat cultivar | External | Internal |
| Alixan | 6.91 ± 2.53 | 5.41 ± 2.48 |
| Altigo | 3.36 ± 1.23 | 1.18 ± 0.40 |
| Cellule | 0.59 ± 0.42 | 18.6 ± 3.94 |
| Hyfi | 1.5 ± 0.73 | 3.61 ± 0.38 |
Table 4: Paenibacillus sp. strain B2 (PB2) external and internal wheat root colonisation increasing ratios in roots inoculated with Mix-3 (PB2: SSM001: SSM004) compared to that inoculated only with PB2. The values shown are the means with SD (n=5).
Impact of wheat genotype on root colonisation by Mix-3
Internal and external root colonisation was evaluated 3 weeks after the inoculation of wheat pregerminated grains at sowing. Each of the three PGPR composing Mix-3 was determined by qPCR using 16S rDNA-specific primers. The results in Figure 2 shows the low external and internal root colonisation with PB2 (≤ 276 pg/g and ≤ 87 pg/g, respectively), compared to SSM-001 (>2.2 × 104 and 107 pg/g) and SSM-004 (1.9 × 104 and 891 pg/g). These root colonisation levels were significantly different than those of PB2 in the four tested cultivars concerning external colonisation and in Altigo and Hyfi for internal colonisation. Furthermore, significant variations in external root colonisation with PB2 were observed between cultivars, where less colonisation was observed in Cellule compared to Alixan and Altigo. On the other hand, internal root colonisation with SSM-001 and SSM-004 was significantly higher in Hyfi than in the other cultivars (Figure 2).
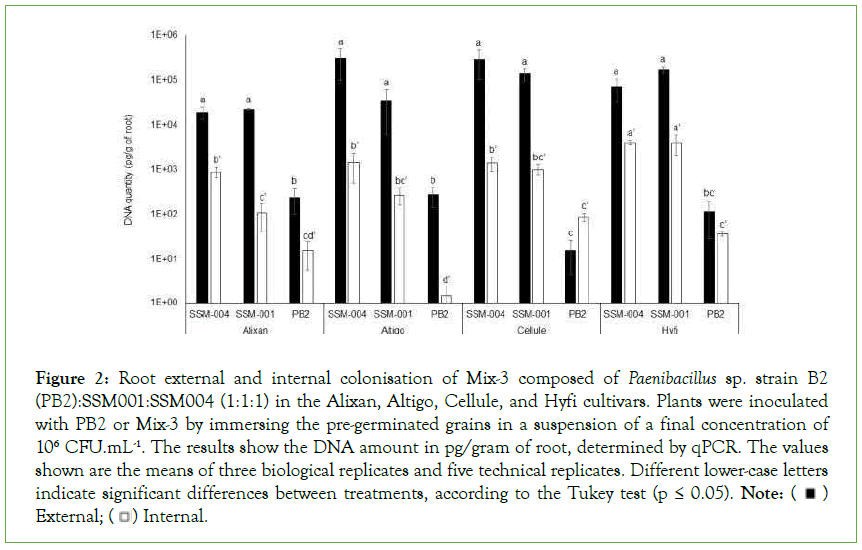
Figure 2: Root external and internal colonisation of Mix-3 composed of Paenibacillus sp. strain B2
(PB2):SSM001:SSM004 (1:1:1) in the Alixan, Altigo, Cellule, and Hyfi cultivars. Plants were inoculated
with PB2 or Mix-3 by immersing the pre-germinated grains in a suspension of a final concentration of
106 CFU.mL-1. The results show the DNA amount in pg/gram of root, determined by qPCR. The values
shown are the means of three biological replicates and five technical replicates. Different lower-case letters
indicate significant differences between treatments, according to the Tukey test (p ≤ 0.05). Note: External;
External; Internal.
Internal.
Durability of root colonisation by Mix-3
The durability of root colonisation with the three PGPR that composed Mix-3 was evaluated at F-L GS in the Alixan and Cellule cultivars using qPCR. However, the three PGPR were present and no significant differences were observed between the two tested cultivars. High levels of external (2.7 × 106 and 5 × 105 pg/g) and internal (1 × 103 and 2.4 × 105 pg/g) root colonisation with SSM- 001 were observed in the Cellule and Alixan cultivars, respectively, followed by SSM-004 with (3.7 × 103 and 2.9 × 103 pg/g) and (8 × 10 and 5.1 × 102 pg/g) and PB2 with (30 and 55 pg/g) and (10 and 7 pg/g) for external and internal root colonisation in the Cellule and Alixan cultivars, respectively (Figure 3).
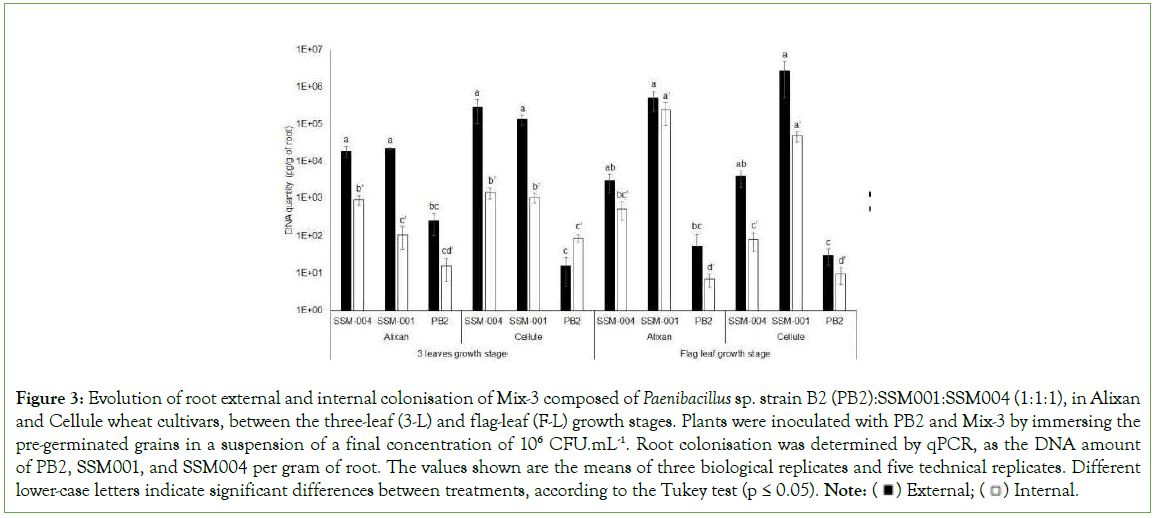
Figure 3: Evolution of root external and internal colonisation of Mix-3 composed of Paenibacillus sp. strain B2 (PB2):SSM001:SSM004 (1:1:1), in Alixan and Cellule wheat cultivars, between the three-leaf (3-L) and flag-leaf (F-L) growth stages. Plants were inoculated with PB2 and Mix-3 by immersing the
pre-germinated grains in a suspension of a final concentration of 106 CFU.mL-1. Root colonisation was determined by qPCR, as the DNA amount
of PB2, SSM001, and SSM004 per gram of root. The values shown are the means of three biological replicates and five technical replicates. Different lower-case letters indicate significant differences between treatments, according to the Tukey test (p ≤ 0.05). Note: External;
External; Internal.
Internal.
Impact of Mix-3 on plant growth promotion
The impact of Mix-3 on FDB and RDB was evaluated 6 weeks after sowing in the four cultivars, i.e., Alixan, Altigo, Cellule and Hyfi, and the results were compared with the impact of PB2 in single inoculation modalities. PB2 did not demonstrate any beneficial or negative effect on root and foliar biomass of wheat. Mix-3, composed of PB2, SSM-001, and SSM-004, showed a significant increase in FDB of 48% in Altigo and an increase of 16% and 10% in Cellule and Hyfi, respectively. Root biomass was also doubled by Mix-3 in Alixan and Altigo and showed an increase of 62% in Cellule compared to the non-inoculated control. In Hyfi, Mix-3 maintained the same root biomass as control (Figure 4).
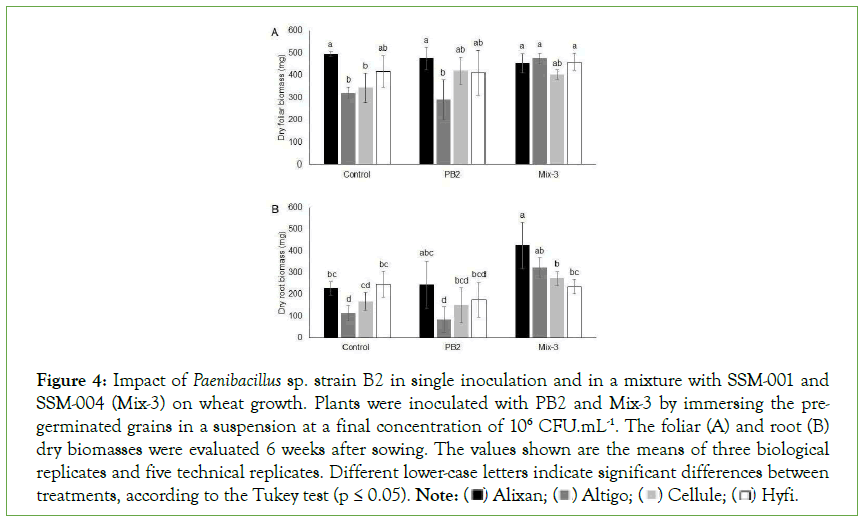
Figure 4: Impact of Paenibacillus sp. strain B2 in single inoculation and in a mixture with SSM-001 and SSM-004 (Mix-3) on wheat growth. Plants were inoculated with PB2 and Mix-3 by immersing the pregerminated
grains in a suspension at a final concentration of 106 CFU.mL-1. The foliar (A) and root (B)
dry biomasses were evaluated 6 weeks after sowing. The values shown are the means of three biological
replicates and five technical replicates. Different lower-case letters indicate significant differences between treatments, according to the Tukey test (p ≤ 0.05). Note: Hyfi.
Hyfi.
Resistance induction by Mix-3 in wheat against drought stress
The Drought Induced Tolerance (DIT) by PB2 and Mix-3 was evaluated using the four wheat cultivars, Alixan, Altigo, Cellule, and Hyfi, after 19 days of drought stress without watering followed by two weeks of a recovery period with normal watering.
The results showed a significant decrease, in all tested cultivars, for foliar and root dry biomasses as a response to drought stress. A considerable impact was observed in Altigo compared to the three other cultivars, and it failed to recover growth after watering. The results in Figure 5 shows a significant reduction in wheat root length in response to drought stress compared to non-stressed controls. The dry foliar biomass results demonstrate the protective effect of PB2 in three wheat cultivars, i.e., Alixan, Altigo, and Cellule, with 56%, 65%, and 42%, respectively, compared to the stressed-noninoculated controls. The dry foliar biomass results showed also significant protection efficiency, but only in two cultivars, Alixan and Cellule, with 65% and 63%, respectively, compared to stressed, non-inoculated controls (Figure 5).
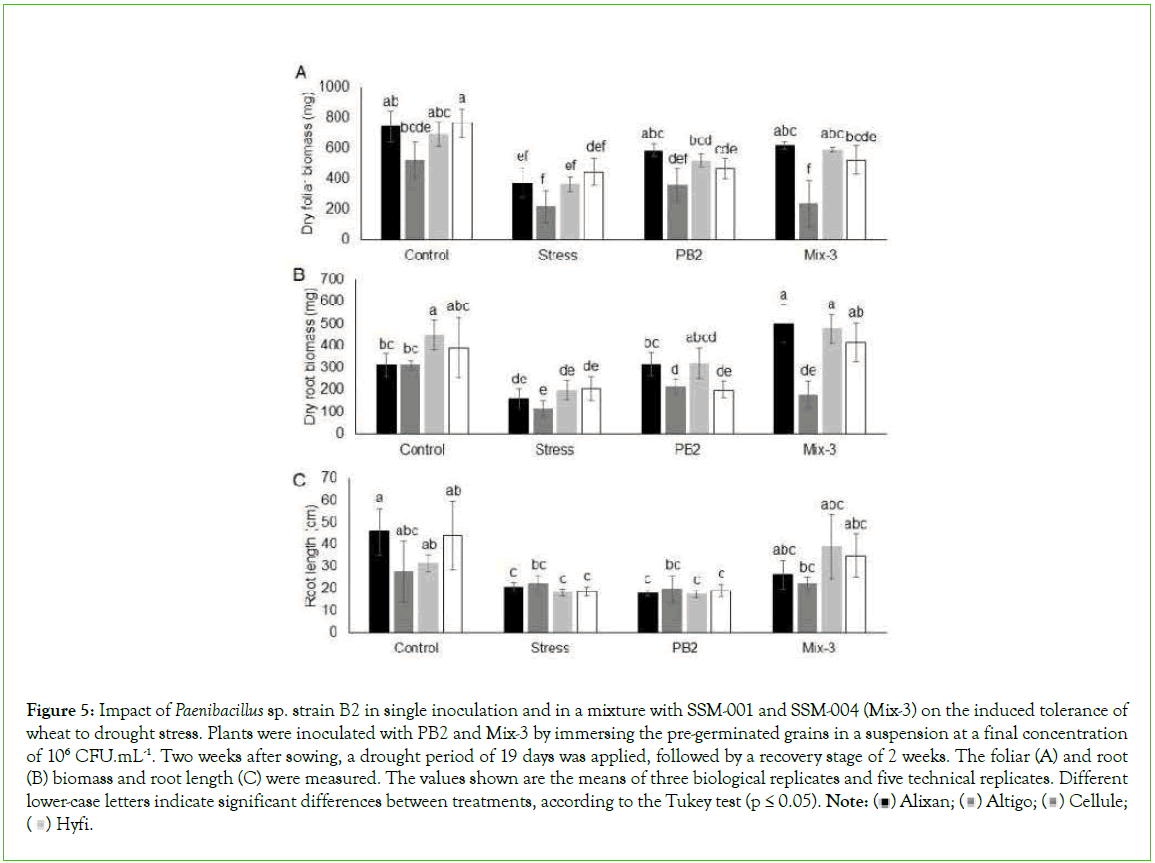
Figure 5: Impact of Paenibacillus sp. strain B2 in single inoculation and in a mixture with SSM-001 and SSM-004 (Mix-3) on the induced tolerance of
wheat to drought stress. Plants were inoculated with PB2 and Mix-3 by immersing the pre-germinated grains in a suspension at a final concentration
of 106 CFU.mL-1. Two weeks after sowing, a drought period of 19 days was applied, followed by a recovery stage of 2 weeks. The foliar (A) and root
(B) biomass and root length (C) were measured. The values shown are the means of three biological replicates and five technical replicates. Different lower-case letters indicate significant differences between treatments, according to the Tukey test (p ≤ 0.05). Note: Hyfi.
Hyfi.
The dry root biomass results showed increases of 214%, 57%, 142%, and 102% with Mix-3 modalities and 98%, 88%, 63%, and 0% with PB2 modalities compared to stressed, non-inoculated controls for Alixan, Altigo, Cellule, and Hyfi, respectively (Figure 5).
Concerning root length, the PB2 modalities did not show significant differences compared to stressed, non-inoculated plants in the four tested cultivars. However, the significant reduction in root length observed in the water stressed modalities was eliminated in the four tested cultivars in the Mix-3 modalities (Figure 5).
Mix-3-induced resistance in wheat against M. graminicola/ impact of wheat genotype on the resistance induced by Mix-3
The protective effect of PGPR Mix-3 was evaluated using the four tested wheat cultivars, Alixan, Altigo, Cellule, and Hyfi, and two M. graminicola strains, IPO323 and 1193 at the 3-L GS compared to PB2 in a single inoculation. The infection levels in controls infected with M. graminicola and non-inoculated with PGPR (M. graminicola- infected-PGPR-non-inoculated controls) were 10903, 376, 396 and 4444 BCN100 ng respectively in the Alixan, Altigo, Cellule, and Hyfi cultivars for strain IPO323 and 511, 770, 189, and 217 BCN100 ng, respectively, for strain 1193. However, these infection levels were strongly reduced in response to root inoculation with Mix-3 in the four tested cultivars, with 76%-89% protection against strain IPO323 and 74%-83% against strain 1193, compared to 58%-75% and 59%-95%, respectively, in response to PB2 (Figure 6).
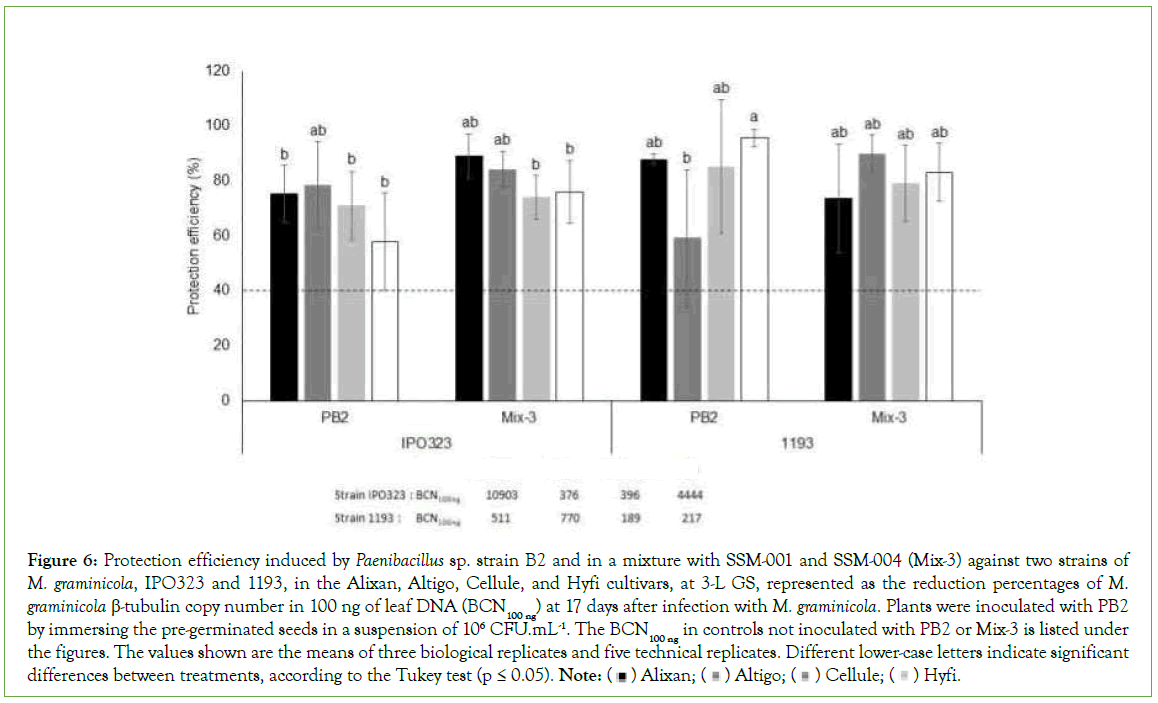
Figure 6: Protection efficiency induced by Paenibacillus sp. strain B2 and in a mixture with SSM-001 and SSM-004 (Mix-3) against two strains of M. graminicola, IPO323 and 1193, in the Alixan, Altigo, Cellule, and Hyfi cultivars, at 3-L GS, represented as the reduction percentages of M.
graminicola β-tubulin copy number in 100 ng of leaf DNA (BCN100 ng) at 17 days after infection with M. graminicola. Plants were inoculated with PB2 by immersing the pre-germinated seeds in a suspension of 106 CFU.mL-1. The BCN100 ng in controls not inoculated with PB2 or Mix-3 is listed under the figures. The values shown are the means of three biological replicates and five technical replicates. Different lower-case letters indicate significant
differences between treatments, according to the Tukey test (p ≤ 0.05). Note: Hyfi.
Hyfi.
The impact of Mix-3 on the pathogen sporulation was studied using epifluorescence microscopy at 21 days after inoculation with the M. graminicola GFP-recombinant strain IPO323. The most susceptible and moderately resistant cultivars, Alixan and Cellule, respectively, were used. In the M. graminicola-infected PGPR-non- inoculated controls, the leaf necrotised area with pycnidia was 25% and 7.75% for Alixan and Cellule, respectively. As a response to root inoculation with Mix-3, the necrotised lesions with pycnidia were significantly decreased by 68% and 87% for the Alixan and Cellule cultivars, respectively (Figure 7).
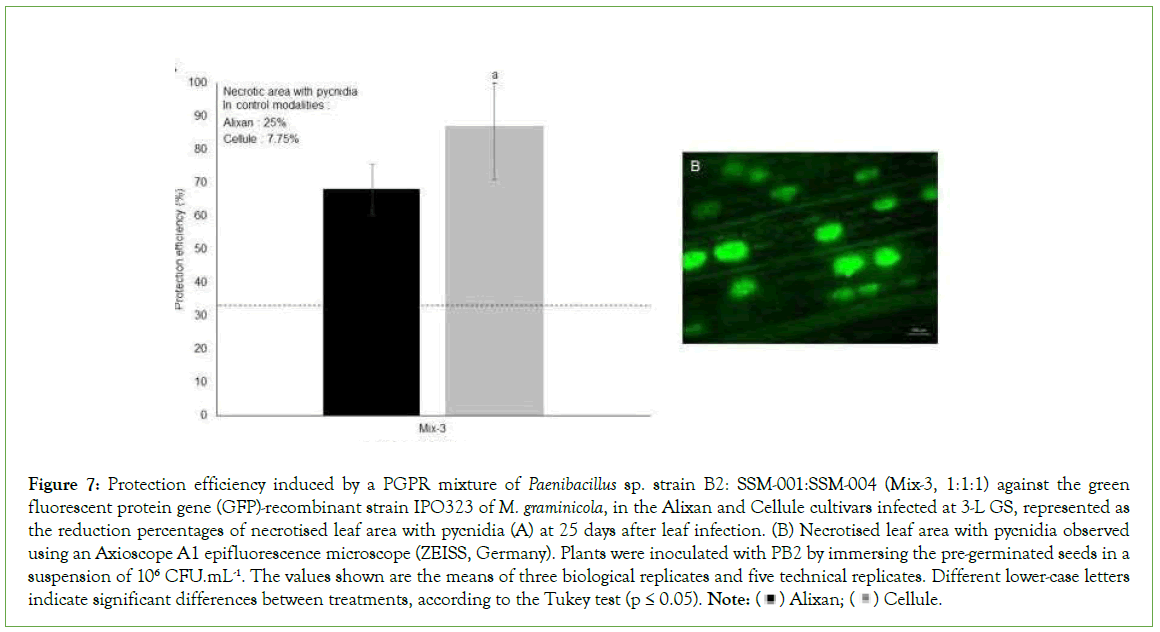
Figure 7: Protection efficiency induced by a PGPR mixture of Paenibacillus sp. strain B2: SSM-001:SSM-004 (Mix-3, 1:1:1) against the green
fluorescent protein gene (GFP)-recombinant strain IPO323 of M. graminicola, in the Alixan and Cellule cultivars infected at 3-L GS, represented as
the reduction percentages of necrotised leaf area with pycnidia (A) at 25 days after leaf infection. (B) Necrotised leaf area with pycnidia observed
using an Axioscope A1 epifluorescence microscope (ZEISS, Germany). Plants were inoculated with PB2 by immersing the pre-germinated seeds in a
suspension of 106 CFU.mL-1. The values shown are the means of three biological replicates and five technical replicates. Different lower-case letters
indicate significant differences between treatments, according to the Tukey test (p ≤ 0.05). Note:  Cellule.
Cellule.
Impact of wheat-genotype growth stage/M. graminicola strain interactions on the durability of resistance induced by Mix-3
The durability of the resistance induced by Mix-3 was evaluated over three wheat growth stages (3-L, Ti, and FL) of the Alixan and Cellule cultivars and against four pathogen strains (IPO323, 1193, TO256, and ST38).
The results in Figure 8 shows strong and stable protection efficiency (>60%) against strain IPO323 on the two tested cultivars and at the three tested growth stages as a response to root inoculation with Mix-3. The protective effect of Mix-3 against strain TO256 was more than 53% and also stable over all tested growth stages in cultivar Cellule, but only in the most mature stages (Ti and F-L GS) for Alixan. Against strain 1193, Mix-3 induced a non-genotype and growth stage-dependent resistance, where the same impact on the two tested cultivars with >48% of protection at 3-L and F-L GS and no protection at Ti GS were observed. For the ST38 strain, Mix-3 was wheat genotype and growth stage dependent, where strong and stable protection (>60%) was observed over the three tested growth stages in Alixan, but only at F-L GS for Cellule (Figure 8).
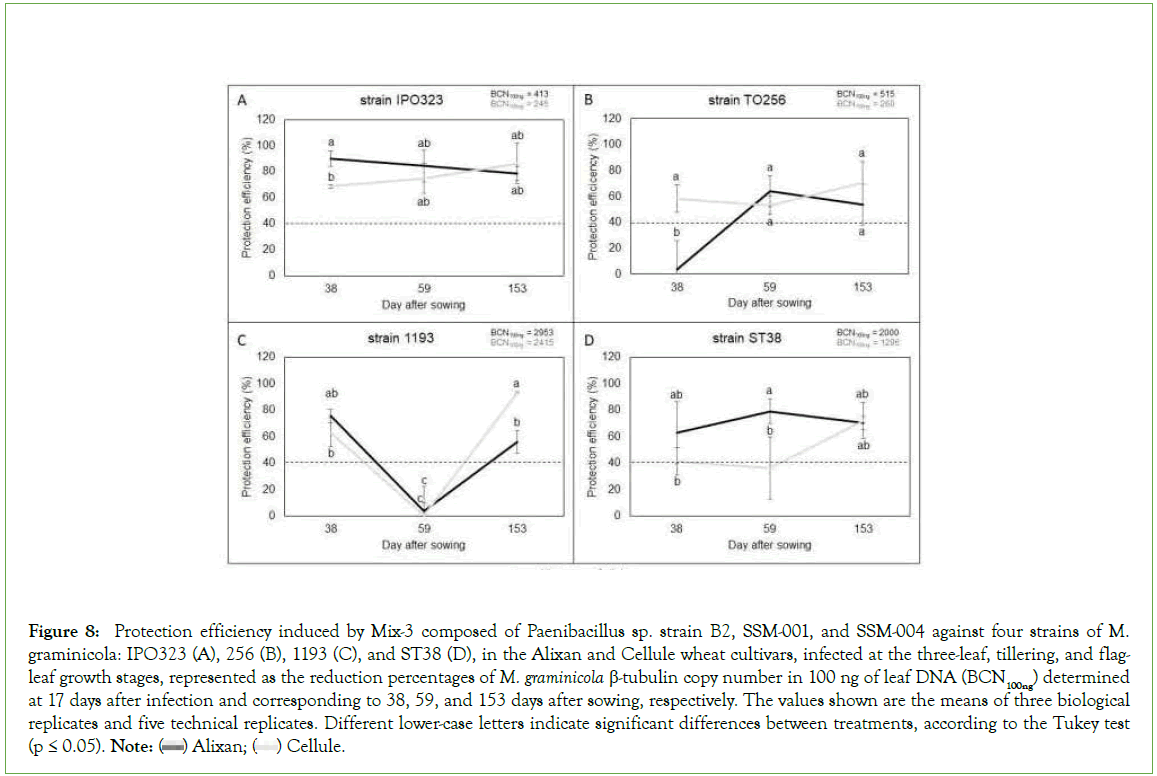
Figure 8: Protection efficiency induced by Mix-3 composed of Paenibacillus sp. strain B2, SSM-001, and SSM-004 against four strains of M. graminicola: IPO323 (A), 256 (B), 1193 (C), and ST38 (D), in the Alixan and Cellule wheat cultivars, infected at the three-leaf, tillering, and flagleaf
growth stages, represented as the reduction percentages of M. graminicola β-tubulin copy number in 100 ng of leaf DNA (BCN100ng) determined at 17 days after infection and corresponding to 38, 59, and 153 days after sowing, respectively. The values shown are the means of three biological
replicates and five technical replicates. Different lower-case letters indicate significant differences between treatments, according to the Tukey test (p ≤ 0.05). Note:  Cellule.
Cellule.
Gene expression time-course analysis
To better understand the defence mechanisms implicated in the wheat genotype/M. graminicola-Mix-3 interaction, the expression of twenty defence-related genes in wheat leaves was studied, using three-week-old plants of the Alixan and Cellule cultivars, as a response to root inoculation with Mix-3 and/or leaf infection with M. graminicola strain IPO323, at T0 (at the time of leaf infection), 6, 12, 24, and 48 hai and at 3, 5, 9, and 11 dai (Figures 9 and 10) and (Supplementary Table 2).
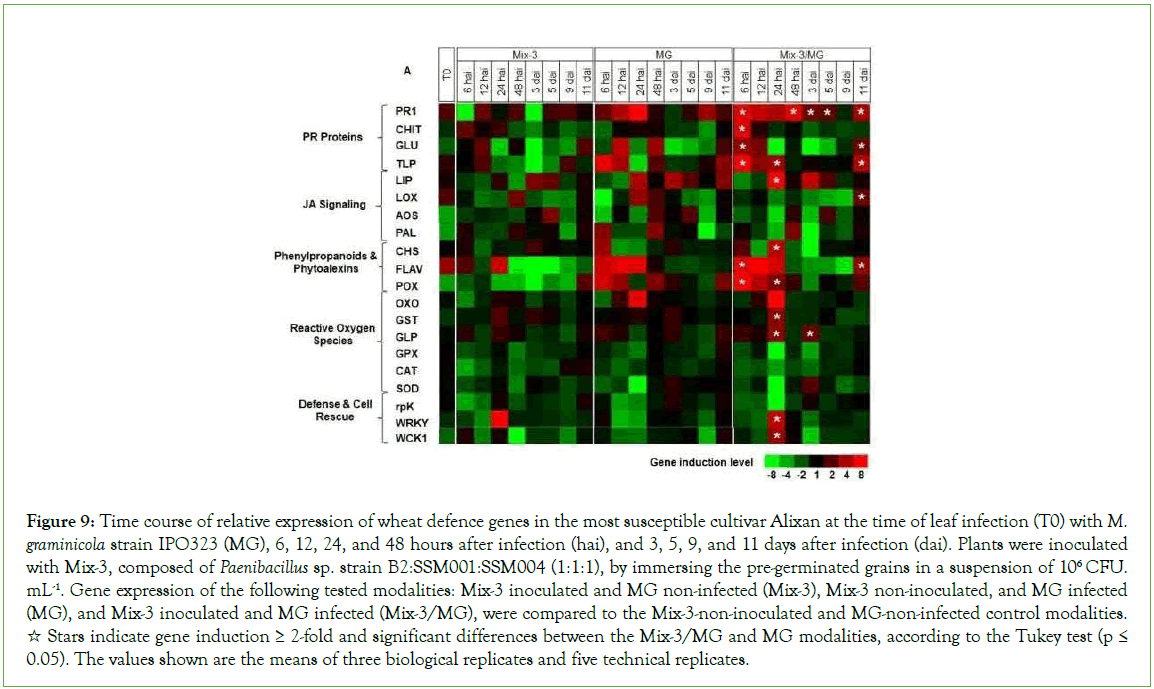
Figure 9: Time course of relative expression of wheat defence genes in the most susceptible cultivar Alixan at the time of leaf infection (T0) with M. graminicola strain IPO323 (MG), 6, 12, 24, and 48 hours after infection (hai), and 3, 5, 9, and 11 days after infection (dai). Plants were inoculated with Mix-3, composed of Paenibacillus sp. strain B2:SSM001:SSM004 (1:1:1), by immersing the pre-germinated grains in a suspension of 106 CFU. mL-1. Gene expression of the following tested modalities: Mix-3 inoculated and MG non-infected (Mix-3), Mix-3 non-inoculated, and MG infected (MG), and Mix-3 inoculated and MG infected (Mix-3/MG), were compared to the Mix-3-non-inoculated and MG-non-infected control modalities. ☆ Stars indicate gene induction ≥ 2-fold and significant differences between the Mix-3/MG and MG modalities, according to the Tukey test (p ≤ 0.05). The values shown are the means of three biological replicates and five technical replicates.
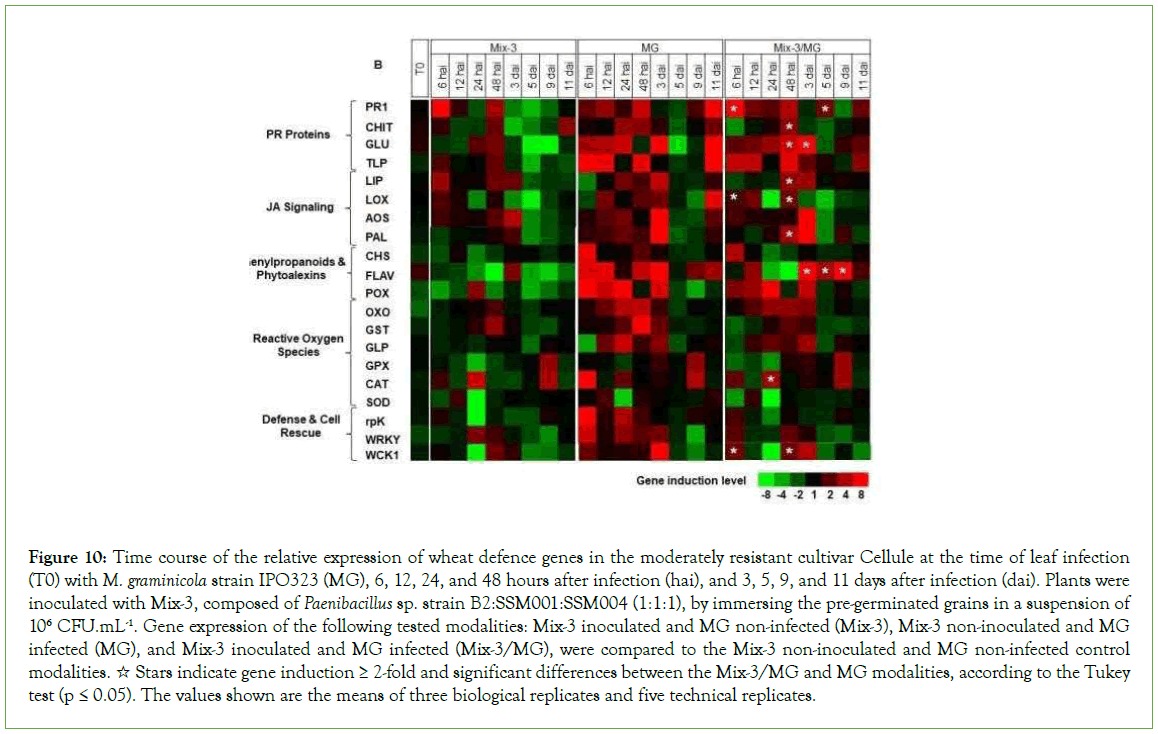
Figure 10: Time course of the relative expression of wheat defence genes in the moderately resistant cultivar Cellule at the time of leaf infection (T0) with M. graminicola strain IPO323 (MG), 6, 12, 24, and 48 hours after infection (hai), and 3, 5, 9, and 11 days after infection (dai). Plants were inoculated with Mix-3, composed of Paenibacillus sp. strain B2:SSM001:SSM004 (1:1:1), by immersing the pre-germinated grains in a suspension of 106 CFU.mL-1. Gene expression of the following tested modalities: Mix-3 inoculated and MG non-infected (Mix-3), Mix-3 non-inoculated and MG infected (MG), and Mix-3 inoculated and MG infected (Mix-3/MG), were compared to the Mix-3 non-inoculated and MG non-infected control modalities. ☆ Stars indicate gene induction ≥ 2-fold and significant differences between the Mix-3/MG and MG modalities, according to the Tukey test (p ≤ 0.05). The values shown are the means of three biological replicates and five technical replicates.
In the absence of pathogen, at T0, significant upregulation was observed in leaves of the cultivar Alixan for the PR1, LOX, and FLAV genes by 2.2, 1.9, and 3.5-fold, respectively, and in Cellule for FLAV by 1.8-fold.
In the presence of pathogen, genes that showed more significant upregulation in the Mix-3/MG modalities than in the MG modalities are labelled with stars in Figures 9 and 10. These genes were used to discriminate the effect of Mix-3 from that of MG strains and to illustrate the priming impact of Mix-3. In Alixan, the results in Figure 9 and Supplementary Table 2 shows significant overexpression of genes implicated in basal defences (PR1, CHIT, GLU, TLP, and LIP) with average inductions, over all tested time points, by 4.9, 7, 3.1, 2.6, and 15-fold, respectively; defence and cell rescue (WRKY1 and WCK1) by 6.1 and 5.6-fold, respectively; the JA signalling pathway (LOX) by 2.3-fold, the phytoalexin and phenylpropanoid pathway (CHS and FLAV) by 22 and 5.7-fold, respectively; and ROS pathway (POX, GST, and GLP) by 6.1, 3.5 and 3.3-fold, respectively. Almost of them were upregulated early at 6 to 24 hai except LOX at 11 dai, FLAV from 48 hai to 9 dai, POX at 3, 5, and 9 dai, and GLP at 48 hai, 5, 9, and 11 dai.
In Cellule, the results in Figure 10 and Supplementary Table 2 shows significant upregulation of genes implicated in basal defences (PR1, CHIT, GLU and LIP) with average inductions, over all tested time points, of 14.3, 3.8, 12.4, and 3.5-fold, respectively; defence and cell rescue (WCK1) by 3.6-fold; the JA signalling pathway (LOX) by 2.4-fold; the phytoalexin and phenylpropanoid pathway (PAL and FLAV) by 4.1 and 18.7-fold, respectively, and the ROS pathway (OXO and CAT) by 3.1 and 4.3-fold, respectively. Also, almost of them were upregulated early, within hours after inoculation, except for PR1 which was also upregulated at 5 and 11 dai, FLAV at 5, 9, and 11 dai, and CAT at 9 dai.
Discussion
We previously showed the importance of plant genotypes on external and internal plant root colonisation with PGPR, especially when applied as single strains [2]. Plant resistance induced against STB is also influenced by plant growth stage and pathogen strain, according to Samain et al. In the present study, we aimed to evaluate the efficiency of a mixture of PGPR on wheat protection and growth promotion, taking into consideration plant genotype, growth stage, and pathogen strains.
The results of the first screening of PGPR showed considerable variations in the response of wheat cultivars to root colonisation, especially internally, as well as in the Induced Resistance (IR) against STB and plant growth promotion. Moreover, some of the screened PGPR showed resistance induction against STB with the inhibition of plant growth. This observation can be explained by the energetic cost of ISR in the plant, broadly described in the literature [18,19]. However, PGPR enhanced plant growth and/or ISR were selected, and a mixture of the three best PGPR was developed from PB2, SSM-001, and SSM-004, referred to in this study as Mix-3.
As observed previously by Samain et al., we confirmed, using more wheat cultivars, that PB2 alone has no impact on wheat growth promotion. However, PB2 in co-inoculation with SSM-001 and SSM-004 in Mix-3 showed a significant increase in root biomass in the Alixan, Altigo, and Cellule cultivars and significantly promoted foliar dry biomass in Altigo. These results indicate the synergistic beneficial impact of combining the PGPR SSM001 and SSM004 with PB2, even though their growth promotion impact was cultivar-dependent. Several studies have shown greater effectiveness of PGPR mixtures compared to single strain inoculations because of the association of multiples mode of action involved, which results synergistic mechanisms [12,20]. Previously, Liu et al. demonstrated that PGPR mixtures containing strains known as resistance inducers and others as plant growth promoters reduce black rot incidence in Chinese cabbage and stimulate plant growth [21].
Here, Mix-3 showed good potential for external and internal root colonisation, more so for SSM-001 and SSM-004 compared to PB2, which seems less competitive compared to the other PGPR on the tested cultivars. Despite these proportional differences between the three PGPR, the PB2 external and internal colonisation levels were increased in co-inoculation compared to single inoculation in almost tested cultivars. These results demonstrate the compatibility of these three PGPR in co-inoculation and the helpful impact of SSM-001 and SSM-004 toward PB2. Interestingly, it has been shown previously that PB2 has also a helping effect on the PGPR Curtobacterium plantarum [2], which indicates more possible advantages when Mix-3 comes into contact with other grain- associated or agricultural soil PGPR.
Under drought stress conditions, all four tested cultivars were influenced, showing significant reductions in foliar and root dry biomass and a tendency to have shorter roots keeping in touch with the soil substrate in pots. However, a severe impact was observed in Altigo compared to the three other cultivars, which made it difficult to recover growth after resuming normal irrigation. However, for the other cultivars, Mix-3 was more efficient than PB2 in terms of protecting plants from weight loss as a response to drought stress. Interestingly, it also maintained root length similar to those of the controls without drought stress. PB2 also reduced the impact of abiotic stress on dry foliar and root biomass, but less effectively than Mix-3 and with strong cultivar dependence, and without any protective effect on root length. As PB2 alone is not Plant Growth Promotor (PGP) in non-stressed conditions, these results suggest that PB2 is PGP stress-dependent, as previously demonstrated for other PGPR such as Acinetobacter and Pseudomonas [22].
However, Mix-3 protected wheat against drought stress in the four wheat cultivars for root length, and in three cultivars for dry root and foliar biomasses; there was even an increase in Alixan root biomass compared to the non-stressed control. The enhancement of root growth under drought stress using Mix-3 compared to the single inoculation of PB2 might be explained by a complementary effect of PGPR enhancing growth under non-stressed conditions [11,23]. Interestingly, Altigo, the only cultivar without tolerance induced against water stress in the presence of Mix-3, was also the only cultivar where the internal root colonisation with PB2 was not increased in Mix-3 compared to single inoculation. These results may suggest the importance of PB2 internal root colonisation in induced tolerance against water stress and the importance of the helping effect of the two other PGPR in Mix-3. Indeed, previous works have demonstrated that endophytic bacteria stimulate plant defence mechanisms against abiotic stress and enhance plant growth under drought [24,25] by inducing antioxidant defence systems to prevent oxidative damage in plants occurring under water stress via the up-regulation of genes such as Super-Oxide Dismutase (SOD), Catalase (CAT), or Glutathione Peroxidase (GPX) [26,27]. The FLAV gene, coding for flavonoid 7-O-methyltransferase implied in flavonoid methylation and phytoalexin synthesis [28], was demonstrated by Ma et al. to be an important defence gene in wheat tolerance to drought stress [29]. Furthermore, FLAV was the only gene overexpressed in both cultivars under non-stressed conditions, as a response to wheat root inoculation with Mix-3 at sowing, which may explain the IR against drought stress observed in this study.
On the other hand, compared to PB2 in single inoculation, Mix- 3 demonstrated stronger and more stable protection efficiency against STB, at 3-L GS, on the four tested cultivars. This protection efficiency as a response to Mix-3 was durable and protected the flag leaf, the most important leaf layer for grain filling, and against the four tested M. graminicola strains. These results confirm the greater efficacy, durability, and stability through pathogen strains and plant genotypes of PGPR in the mixture compared to individual strains [30].
In the absence of pathogen, the gene expression results show that only PR1, LOX, and FLAV were up-regulated in Alixan, and only FLAV in Cellule as a response to root inoculation with Mix-3. A priming effect was observed in almost all tested genes for Mix-3 modalities after leaf infection with M. graminicola. Significant upregulation compared to the Mix-3 non-inoculated and M. graminicola-infected modalities were observed for the genes PR1, CHIT, GLU, LIP, LOX, WCK1, and FLAV in the two tested cultivars, and in addition to TLP, CHS, POX, GST, GLP, and WRKY1 for Alixan, and PAL and CAT for Cellule. The antagonistic properties against M. graminicola of PR proteins such as CHIT (PR-3) and GLU (PR-2) are now well-known as coding for the degrading enzymes of fungal cell walls and contributing to the state of resistance in wheat [2,7,16,31-33]. The overexpression of CHIT was previously observed in resistant wheat cultivars against M. graminicola [34] and might have protective efficiency against Phaeosphaeria nodorum in wheat in combination with GLU [35]. Furthermore, GLU, combined with TLP (PR-5), as in this work, in the susceptible cultivar Alixan, already demonstrated a synergistic effect against M. graminicola [36]. Indeed, this work shows the similarities in the timing of upregulation between these genes at 6 and 12 hai (CHIT, GLU and TLP) and 11 dai (GLU and TLP) in Alixan and at 48 hai (CHIT, GLU and TLP) and 6, 12, and 24 hai, and 11 dai (GLU and TLP) in Cellule, which suggests a common overexpression signal. However, GLU and CHIT are also implied in the propagation of wheat defence signals, through the release of fungal chitin and β-1,3-glucan fragments (PAMPs, (Pathogen-Associated Molecular Patterns)) in the intracellular space [33]. One of the most prolonged overexpressed genes is PR1, from 6 hai to 11 dai, except for 9 dai in Alixan and 3 and 9 dai in Cellule, even in the presence of pathogen, suggesting once again its importance in plant defence mechanisms against M. graminicola, during both the biotrophic and necrotrophic stages [2,7,31]. Additionally, the overexpression of LIP, coding for lipase, is known to disrupt invading pathogens and acts on Arabidopsis thaliana defence pathways through the generation of lipid-derived molecules, synthetised by the hydrolysing properties of the enzyme. These molecules are suggested to be lipid transfer protein (PR-14) forms [37]. LIP seems only intervene at late stage of pathogen development, i.e. from 24 hai and 48 hai for Alixan and Cellule, respectively. LOX and WCK1, both induced by the JA signalling pathway, were overexpressed in Alixan and Cellule in response to Mix-3 after M. graminicola infection. LOX catalyses the deoxygenation of polyunsaturated fatty acids and belongs to the octadecanoid pathway. As demonstrated by Somai-Jemmali et al., our results show LOX down-regulation at 24 hai in the resistant and the susceptible cultivars, and upregulation at 6, 12, and 48 hai in Cellule and at 11 dai in Alixan. WCK1 (the mitogen-activated protein kinase (MAPK) of wheat) is only upregulated in Alixan at 24 hai and in Cellule at 6 and 48 hai, and at 3 dai. MAPK are known as plant defence reaction inducers against biotic and abiotic stresses and may be associated with the transcription factor WRKY1 [38-42]; they are generally expressed early but, in the case of M. graminicola infection, expression is delayed, probably due to the late trigger of the necrotrophic phase of the pathogen [43-45]. The upregulation of FLAV, belonging to the phenylpropanoid and phytoalexin pathway, supplemented by CHS in Alixan and PAL in Cellule, and the overexpression of POX, GST, GLP and CAT, result in ROS pathway activation confirm our previous results as a response to PB2 in a single inoculation [2,7,46].
Conclusion
Paenibacillus sp. strain B2 is a potential biological control agent against Septoria tritici leaf blotch and drought stress. Its efficiency was improved by co-inoculation with the PGPR SSM-001 and SSM- 004, exhibiting higher, more stable, and more durable protection. Moreover, the impact of wheat genotype and pathogen strain, observed in a single inoculation, was reduced. Mix-3 also promoted plant growth with or in the absence of stress conditions.
Our results highlight the importance of basal defences, ROS, phenylpropanoids and phytoalexins, and the SA and JA pathways in the resistance induced in wheat by Mix-3, with importance contribution of PR1, chitinase, glucanase and flavonoides to the resistance of wheat to M. graminicola and drought stresses.
Author Contribution
E.S. carried out the experimental work and wrote the manuscript. S.S. and T.A. revised the manuscript, and managed the experimental design and the PhD research program. S.S., T.A. and C.E. supervised the project.
Funding Statement
This work was supported by SDP society and ANRT “Association Nationale de la Recherche et de la Technologie” under convention N° 56/2016.
Acknowledgement
We are grateful to SDP for financing this work, with the support of the French ‘Association National Recherche Technologie (ANRT)’, under convention N° 56/2016. We would like to thank the excellent contribution of the SDP department of Research, Innovation, and Technics.
REFERENCES
- Kundan R, Pant G, Jadon N, Agrawal PK. Plant growth promoting Rhizobacteria: Mechanism and current prospective. J Fertil Pestic. 2015;6(2):9.
- Samain E, van Tuinen D, Jeandet P, Aussenac T, Selim S. Biological control of septoria leaf blotch and growth promotion in wheat by Paenibacillus sp. strain B2 and Curtobacterium plantarum strain EDS. Biol Cont. 2017;114:87-96.
- Sarma BK, Yadav SK, Singh S, Singh HB. Microbial consortium-mediated plant defense against phytopathogens: Readdressing for enhancing efficacy. Soil Biol Biochem. 2015;87:25-33.
- Selim S, Negrel J, Wendehenne D, Ochatt S, Gianinazzi S, van Tuinen D. Stimulation of defense reactions in Medicago truncatula by antagonistic lipopeptides from Paenibacillus sp. strain B2. Appl Environ Microbiol. 2010;76(22):7420-7428.
- Morales-García YE, Juárez-Hernández D, Aragón-Hernández C, Mascarua-Esparza MA, Bustillos-Cristales MR, Fuentes-Ramírez LE, et al. Growth response of maize plantlets inoculated with Enterobacter spp., as a model for alternative agriculture. Argent J Microbiol. 2011;43(4):287-293.
[Crossref] [Google Scholar] [PubMed]
- Molinari S. Natural genetic and induced plant resistance, as a control strategy to plant-parasitic nematodes alternative to pesticides. Plant Cell Rep. 2011;30(3):311-23.
[Crossref] [Google Scholar] [PubMed]
- Samain E, Aussenac T, Selim S. The effect of plant genotype, growth stage, and Mycosphaerella graminicola strains on the efficiency and durability of wheat-induced resistance by Paenibacillus sp. Strain B2. Front Plant Sci. 2019;10:587.
[Crossref] [Google Scholar] [PubMed]
- Soussi A, Ferjani R, Marasco R, Guesmi A, Cherif H, Rolli E, et al. Plant-associated microbiomes in arid lands: Diversity, ecology and biotechnological potential. Plant Soil. 2016;405(1):357-370.
- Yang J, Kloepper JW, Ryu CM. Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 2009;14(1):1-4.
[Crossref] [Google Scholar] [PubMed]
- Timmusk S, Wagner EG. The plant-growth-promoting Rhizobacterium Paenibacillus polymyxa induces changes in Arabidopsis thaliana gene expression: A possible connection between biotic and abiotic stress responses. Mol Plant Microbe Interact. 1999;12(11):951-959.
[Crossref] [Google Scholar] [PubMed]
- Vurukonda SS, Vardharajula S, Shrivastava M, SkZ A. Enhancement of drought stress tolerance in crops by plant growth promoting Rhizobacteria. Microbiol Res. 2016;184:13-24.
[Crossref] [Google Scholar] [PubMed]
- Raupach GS, Kloepper JW. Mixtures of plant growth-promoting Rhizobacteria enhance biological control of multiple cucumber pathogens. Phytopathol. 1998;88(11):1158-1164.
[Crossref] [Google Scholar] [PubMed]
- Selim S, Negrel J, Govaerts C, Gianinazzi S, Van Tuinen D. Isolation and partial characterization of antagonistic peptides produced by Paenibacillus sp. strain B2 isolated from the sorghum mycorrhizosphere. Appl Environ Microbiol. 2005;71(11):6501-6507.
[Crossref] [Google Scholar] [PubMed]
- Budi SV, van Tuinen D, Martinotti G, Gianinazzi S. Isolation from the Sorghum bicolor mycorrhizosphere of a bacterium compatible with Arbuscular mycorrhiza development and antagonistic towards soil borne fungal pathogens. Appl Environ Microbiol. 1999;65(11):5148-50.
[Crossref] [Google Scholar] [PubMed]
- Selim S, Roisin‐Fichter C, Andry JB, Bogdanow B, Sambou R. Real‐time PCR to study the effect of timing and persistence of fungicide application and wheat varietal resistance on Mycosphaerella graminicola and its sterol 14 α‐demethylation‐inhibitor‐resistant genotypes. Pest Manag Sci. 2014;70(1):60-69.
[Crossref] [Google Scholar] [PubMed]
- Ors ME, Randoux B, Selim S, Siah A, Couleaud G, Maumené C, et al. Cultivar‐dependent partial resistance and associated defence mechanisms in wheat against Zymoseptoria tritici. Plant Pathol. 2018;67(3):561-572.
- Bearchell SJ, Fraaije BA, Shaw MW, Fitt BD. Wheat archive links long-term fungal pathogen population dynamics to air pollution. Proc Natl Acad Sci USA. 2005;102(15):5438-5442.
[Crossref] [Google Scholar] [PubMed]
- Hoffland E, Pieterse CM, Bik L, Van Pelt JA. Induced systemic resistance in radish is not associated with accumulation of pathogenesis-related proteins. Physiol Mol Plant Pathol. 1995;46(4):309-320.
- Heil M, Baldwin IT. Fitness costs of induced resistance: Emerging experimental support for a slippery concept. Trends Plant Sci. 2002;7(2):61-67.
[Crossref] [Google Scholar] [PubMed]
- Marimuthu S, Ramamoorthy V, Samiyappan R, Subbian P. Intercropping system with combined application of Azospirillum and Pseudomonas fluorescens reduces root rot incidence caused by Rhizoctonia bataticola and increases seed cotton yield. J Phytopathol. 2013;161(6):405-411.
- Liu K, Garrett C, Fadamiro H, Kloepper JW. Antagonism of black rot in cabbage by mixtures of Plant Growth-Promoting Rhizobacteria (PGPR). Bio Control. 2016;61(5):605-613.
- Rolli E, Marasco R, Vigani G, Ettoumi B, Mapelli F, Deangelis ML, et al. Improved plant resistance to drought is promoted by the root‐associated microbiome as a water stress‐dependent trait. Environ Microbiol. 2015;17(2):316-331.
[Crossref] [Google Scholar] [PubMed]
- Vardharajula S, Zulfikar Ali S, Grover M, Reddy G, Bandi V. Drought-tolerant plant growth promoting Bacillus spp.: effect on growth, osmolytes, and antioxidant status of maize under drought stress. J Plant Interact. 2011;6(1):1-4.
- Naveed M, Hussain MB, Zahir ZA, Mitter B, Sessitsch A. Drought stress amelioration in wheat through inoculation with Burkholderia phytofirmans strain PsJN. Plant Growth Regulat. 2014;73(2):121-131.
- Chen C, Xin K, Liu H, Cheng J, Shen X, Wang Y, et al. Pantoea alhagi, a novel endophytic bacterium with ability to improve growth and drought tolerance in wheat. Sci Rep. 2017;7(1):1-4..
[Crossref] [Google Scholar] [PubMed]
- Miller GA, Suzuki N, Ciftci‐Yilmaz SU, Mittler RO. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010;33(4):453-467.
[Crossref] [Google Scholar] [PubMed]
- Heidari M, Golpayegani A. Effects of water stress and inoculation with Plant Growth Promoting Rhizobacteria (PGPR) on antioxidant status and photosynthetic pigments in basil (Ocimum basilicum L.). J Saudi Soc Agric Sci. 2012;11(1):57-61.
- Edwards R, Dixon RA. Purification and characterization of s-adenosyl-l-methionine: Caffeic acid 3-O-methyltransferase from suspension cultures of alfalfa (Medicago sativa L.). Arch Biochem Biophys. 1991;287(2):372-379.
[Crossref] [Google Scholar] [PubMed]
- Ma D, Sun D, Wang C, Li Y, Guo T. Expression of flavonoid biosynthesis genes and accumulation of flavonoid in wheat leaves in response to drought stress. Plant Physiol Biochem. 2014;80:60-66.
[Crossref] [Google Scholar] [PubMed]
- Jetiyanon K, Fowler WD, Kloepper JW. Broad-spectrum protection against several pathogens by PGPR mixtures under field conditions in Thailand. Plant Dis. 2003;87(11):1390-1394.
[Crossref] [Google Scholar] [PubMed]
- Adhikari TB, Balaji B, Breeden J, Goodwin SB. Resistance of wheat to Mycosphaerella graminicola involves early and late peaks of gene expression. Physiol Mol Plant Pathol. 2007;71(1-3):55-68.
- Ors M, Randoux B, Siah A, Couleaud G, Maumené C, Sahmer K, et al. A plant nutrient-and microbial protein-based resistance inducer elicits wheat cultivar-dependent resistance against Zymoseptoria tritici. Phytopathology. 2019;109(12):2033-45.
[Crossref] [Google Scholar] [PubMed]
- Shetty NP, Jensen JD, Knudsen A, Finnie C, Geshi N, Blennow A, et al. Effects of β-1, 3-glucan from Septoria tritici on structural defence responses in wheat. J Exp Bot. 2009;60(15):4287-4300.
[Crossref] [Google Scholar] [PubMed]
- Somai-Jemmali L, Siah A, Harbaoui K, Fergaoui S, Randoux B, Magnin-Robert M, et al. Correlation of fungal penetration, CWDE activities and defense-related genes with resistance of durum wheat cultivars to Zymoseptoria tritici. Physiol Mol Plant Pathol. 2017;100:117-125.
- Anand A, Zhou T, Trick HN, Gill BS, Bockus WW, Muthukrishnan S. Greenhouse and field testing of transgenic wheat plants stably expressing genes for thaumatin‐like protein, chitinase and glucanase against Fusarium graminearum. J Exp Bot. 2003;54(384):1101-1111.
[Crossref] [Google Scholar] [PubMed]
- Leah R, Tommerup H, Svendsen IB, Mundy J. Biochemical and molecular characterization of three barley seed proteins with antifungal properties. J Biol Chem. 1991;266(3):1564-1573.
[Crossref] [Google Scholar] [PubMed]
- Oh IS, Park AR, Bae MS, Kwon SJ, Kim YS, Lee JE, et al. Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell. 2005;17(10):2832-2847.
[Crossref] [Google Scholar] [PubMed]
- Ahlfors R, Macioszek V, Rudd J, Brosché M, Schlichting R, Scheel D, et al. Stress hormone‐independent activation and nuclear translocation of mitogen‐activated protein kinases in Arabidopsis thaliana during ozone exposure. Plant J. 2004;40(4):512-22.
[Crossref] [Google Scholar] [PubMed]
- Mayrose M, Bonshtien A, Sessa G. LeMPK3 is a mitogen-activated protein kinase with dual specificity induced during tomato defense and wounding responses. J Biol Chem. 2004;279(15):14819-14827.
[Crossref] [Google Scholar] [PubMed]
- Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated signal transduction kinases frequently occupy target genes. Science. 2006;313(5786):533-536.
[Crossref] [Google Scholar] [PubMed]
- Reyna NS, Yang Y. Molecular analysis of the rice MAP kinase gene family in relation to Magnaporthe grisea infection. Mol Plant Microbe Interact. 2006;19(5):530-540.
[Crossref] [Google Scholar] [PubMed]
- Zhang S, Du H, Klessig DF. Activation of the tobacco SIP kinase by both a cell wall–derived carbohydrate elicitor and purified proteinaceous elicitins from Phytophthora spp. Plant Cell. 1998;10(3):435-449.
[Crossref] [Google Scholar] [PubMed]
- Rudd JJ, Keon J, Hammond-Kosack KE. The wheat mitogen-activated protein kinases TaMPK3 and TaMPK6 are differentially regulated at multiple levels during compatible disease interactions with Mycosphaerella graminicola. Plant Physiol. 2008;147(2):802-815.
[Crossref] [Google Scholar] [PubMed]
- Sameh S, Céline RF, Jean-Baptiste A, Boris B, Thajuddin N. Accuracy of real-time PCR to study Mycosphaerella graminicola epidemic in wheat: From spore arrival to fungicide efficiency. In: Thajuddin N (eds) Fungicides Beneficial and Harmful Aspects. Intech Open, United Kingdom. 2011, pp. 219-238.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402-408.
[Crossref] [Google Scholar] [PubMed]
- Samain E, Ernenwein C, Aussenac Th, Selim S. Effective and durable systemic wheat-induced resistance by a plant-growth-promoting rhizobacteria consortium of Paenibacillus sp. strain B2 and Arthrobacter spp. strain AA against Zymoseptoria tritici and drought stress. Physiol Mol Plant Pathol. 2022;119:101830.
Citation: Samain E, Ernenwein C, Aussenac T, Selim S (2022) Efficacy and Durability of Paenibacillus Sp. Strain B2 in Co-Inoculation with Arthrobacter Sp. SSM-004 and Microbacterium Sp. SSM-001 for Growth Promotion and Resistance Induction in Wheat against Mycosphaerella graminicola and Drought Stress. J Plant Pathol Microbiol. 13:603.
Copyright: © 2022 Samain E, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

