Indexed In
- Open J Gate
- JournalTOCs
- The Global Impact Factor (GIF)
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research - (2019) Volume 7, Issue 2
Association Study between Genes Related to Pharmacokinetics and Pharmacodynamics of Oxycodone and Response to Drug Treatment: A Genetic Cohort Study
Yoshimi A1,2,3#, Yoshijima Y1,4#, Miyazaki M1,2, Kato H1,2, Kato YK1, Yamada K2, Ozaki N3, Kaneko R5, Ishii A5, Mitsuma A6, Sugishita M6, Ando Y6 and Noda Y1,2,3*2Department of Neuropsychopharmacology and Hospital Pharmacy, Nagoya University Graduate School of Medicine, 65 Tsurumai-cho, Showa-ku, Nagoya, Japan
3Department of Psychiatry, Nagoya University Graduate School of Medicine, 65 Tsurumai-cho, Showa-ku, Nagoya, Japan
4Department of Pharmacy, Toyota Memorial Hospital, 1-1 Heiwa-cho, Toyota-shi, Aichi, Japan
5Department of Legal Medicine and Bioethics, Nagoya University Graduate School of Medicine, 65 Tsurumai-cho, Showa-ku, Nagoya, Japan
6Department of Clinical Oncology and Chemotherapy, Nagoya University Graduate School of Medicine, 65 Tsurumai-cho, Showaku, Nagoya, Japan
#Equally contribution
Received: 28-Mar-2019 Published: 30-Apr-2019
Abstract
Objective: Oxycodone is widely used in cancer patients with pain, but interindividual differences in both its analgesic efficacy and adverse effects are major clinical disadvantages to therapeutic use. To explore specific polymorphisms affecting drug plasma concentrations, analgesic efficacy, and adverse effects, we performed an association study between genetic polymorphisms affecting pharmacokinetics and pharmacodynamics of oxycodone and response to drug treatment in cancer patients with pain.
Methods: Blood samples were collected from 50 patients 12 h after administration of oxycodone. Genetic polymorphisms related to the pharmacokinetics and pharmacodynamics of oxycodone [cytochrome P450 (CYP3A4*1G, CYP3A5*3, and CYP2D6*10), P-glycoprotein (ABCB1), and opioid receptor μ1 (OPRM1)] were genotyped by real-time polymerase chain reaction (PCR) and plasma concentrations of oxycodone and noroxycodone were determined by ultra-performance liquid chromatography tandem mass spectrometry (UPLC-MS/MS).
Results: Carriers of specific polymorphisms (CYP3A4 *1G/*1G, CYP3A5 *1/*1, CYP2D6 100CC+CT, and ABCB1 2677TA+TT+AA) were associated with increased average total daily dose of oxycodone. CYP3A4 *1G/*1G and CYP3A5 *1/*1 were also associated with increased number of rescues and plasma concentration of oxycodone. Moreover, OPRM1 118AG+GG carriers were related to greater average total daily dose of oxycodone and increased number of rescues.
Conclusion: These findings suggest that genetic polymorphisms of genes related to pharmacokinetics and pharmacodynamics of oxycodone have a potential impact on clinical responses to drug in cancer patients with pain.
Keywords
Genetic polymorphisms; Cancer; Pharmacokinetic; Pharmacodynamics; Oxycodone; Cytochrome P450; P-glycoprotein; Opioid receptor μ1
Introduction
Oxycodone is a semisynthetic, μ-opioid receptor agonist, which elicits analgesic effects in several pain conditions. It is widely recognized as the primary opioid for cancer pain management. Recently, use of oxycodone has become widespread in Japan following the approval of OxyContin® and/or OXINORM®. Recent guidance by the European Association for Palliative Care on the use of opioids in cancer pain suggests that oxycodone could be used as the first line treatment of moderate to severe cancer pain as an alternative to morphine [1]. Cytochrome P450 (CYP) 3A4 and CYP3A5 catalyze the N-demethylation of orally administered oxycodone to noroxycodone, while CYP2D6 catalyzes it (via O-demethylation) to the circulating metabolite, oxymorphone [2-4]. Noroxycodone exhibits weak antinociceptive potency, and oxymorphone is an active metabolite but is produced at low levels [3,5]. Therefore, oxycodone is likely that primarily mediates both the analgesic and adverse effects in patients being treated. The pharmacokinetics (PK) and pharmacodynamics (PD) of opioids are influenced by several genetic polymorphisms, which can account for part of the observed interindividual variation in both pain relief and adverse effects in patients receiving these medications. Genetic polymorphisms of CYP genes, encoding the main oxycodonemetabolizing enzyme;and P-glycoprotein [adenosine triphosphate (ATP)-binding cassette, sub-family B member 1; ABCB1], encoding an efflux transporter; are the primary polymorphisms that have been associated with the PK of oxycodone [6-13]. Furthermore, an opioid receptor μ1 (OPRM1) polymorphism has been associated with the PD of oxycodone, influencing its analgesic effect [12-18]. Although interindividual differences in both analgesic and adverse response to oxycodone have been reported [19-21], the association between genetic polymorphisms (related to PK/PD of oxycodone) and responses to treatment in Japanese cancer patients remains unclear. Molecular activities related to PK/PD of oxycodone can be modified by several factors including not only genetic polymorphisms, but also environmental factors such as changes in physiological conditions (e.g., age and disease status), and drug-drug or drug-food interactions, and so on. Thus, some factors are related to the culture or customs of the population. These factors may cause interindividual differences in the PK/PD profiles, leading to variations in drug efficacy and/or toxicity. To examine an association between genetic polymorphisms affecting pharmacokinetics and pharmacodynamics of oxycodone efficacy in Japanese cancer patients, we analyzed the association between genetic polymorphisms (CYP3A4, CYP3A5, CYP2D6, ABCB1, and OPRM1), plasma concentrations of oxycodone and its' demethylated metabolites, and analgesic and adverse effects.
Methods
Study population and schedule
Entry criteria were as follows: (1) Japanese cancer patients aged more than 20 years, (2) oxycodone took more than 7 days for cancer pain and baseline oxycodone dosage was not changed for more than 7 days, (3) oxycodone rescue use was less than 3 times. Exclusion criteria were as follows: (1) patients using oxycodone rescue 3 times or more because they were considered to be required baseline titration of oxycodone, (2) patients with severe hepetic injury with Child-Pugh score more than ten, and (3) patients received co-administrated drugs, which potently affect PK/PD of oxycodone. Fifty cancer patients were recruited at Nagoya University Hospital Outpatient Chemotherapy Room. We had performed two interviews with patients on different consultation days. In the first interview, we checked the intensity of pain and adverse effects, and explained to record the pain diary until the next interview (the second interview). In the second interview, we confirmed the intensity of them from the pain diary. Pain intensity of patients receiving oxycodone was determined by observing the daily dose of oxycodone (mg/day), the average number of rescues (immediaterelease oxycodone powder) for breakthrough pain (times/day), the numeric rating scale (NRS: 0=no pain to 10=unbearable pain) from the first and second interviews, and the pain diary. The average total daily dose of oxycodone (mg/day) was calculated from the daily dose and the rescue dose. The adverse effects of oxycodone were determined from the following symptoms after taking oxycodone reported in interviews and the pain diary: the degree of constipation; nausea; and drowsiness were graded according to the National Cancer Institute Common Terminology Criteria for Adverse Events (CTCAE v4.0).
Determination of plasma concentrations of oxycodone and noroxycodone
Plasma concentrations of oxycodone and noroxycodone were simultaneously determined by internal standard method using ultraperformance liquid chromatography tandem mass spectrometry (UPLC-MS/MS).
Sample pretreatment
Blood samples were collected 12 h after administration of oxycodone on the day of the second interview (immediately before next administration of oxycodone). Plasma was separated by centrifugation of next blood samples at 4,000 rpm for 10 min and stored at –80°C until analysis. As an internal standard (IS) solution, 2 μL of 1 ng/mL d6-oxycodone (Cerilliant Co., TX, USA) and 800 μL of acetonitrile (Kanto chemical Co., Inc., Japan) were added to 200 μL of plasma. Samples were mixed for 1 min and then centrifuged at 3,500 rpm for 1 min. After drying and concentration using a centrifugal concentrator (TOMY SEIKO Co., Ltd., Japan), the residue was resuspended in 100 μL of 20 mM ammonium acetate (Kanto chemical Co., Inc.): methanol (Kanto chemical Co., Inc.)=4:1 (v/v%), vortexed, and then transferred to vials ready for UPLC-MS/MS injection.
Standard solution
A series of standard working solutions of oxycodone (Covidien Co., Inc., Ireland) and noroxycodone (Covidien Co., Inc.) were obtained by mixing and diluting the standard stock solution with methanol. Calibration standards were prepared by diluting each standard working solution with blank human plasma (200 μL) to yield the following concentrations: 0, 0.5, 1.0, 2.0, 5.0, 10, 20, 40, 60, and 80 ng/mL for oxycodone and noroxycodone. As an internal standard solution, 1 ng/mL d6-oxycodone (Cerilliant Co.) was added to each of the standards.
UPLC-MS/MS
The UPLC-MS/MS analysis was performed using ACQUITY Ultra Performance LC® (Waters Co., Milford, MA, USA). Separation was achieved using an ACQUITY UPLC® BEH C18 column (100 × 2.1 mm i.d., particle size 1.7 μm;Waters Co.) and the column temperature was maintained at 24°C. The mobile phase consisted of (A) 20 mM ammonium acetate and (B) acetonitrile at a flow rate of 0.5 mL/min. The gradient program was from 18% to 95% B for 2.1 min, held for 2.9 min, ramped from 95% to 18% B in 0.1 min (total runtime of 5.1 min). All changes in mobile phase composition utilized a step gradient. The auto-sampler was maintained at 4°C and the sample injection volume was 10 μL. For the operation in MS/MS mode, electrospray ionization (ESI) with selected reaction monitoring (SRM) was used. The selected ion transitions were divided into four periods, as follows: m/z 316 to m/z 241 for oxycodone, m/z 302 to m/z 187 for noroxycodone, and m/z 322 to m/z 304 for d6-oxycodone. During these analyses, the ESI parameters were set as follows: capillary voltage 3.0 kV for positive mode; ion source temperature 150°C; desolvation temperature 450°C; cone and desolvation gas, nitrogen, with flow rate of with 50 and 800 L/h, respectively. The cone voltages of oxycodone, noroxycodone, and d6-oxycodone were 40, 20, 35, and 30 V, respectively. The collision gas (Argon) flow was 0.14 mL/ min and the collision voltages of oxycodone, noroxycodone and d6-oxycodone were 20, 30, 35, and 25 eV, respectively.
Measurement of oxycodone and noroxycodone in plasma
The results of linear regression analysis showed that correlation coefficients of all standard curves were at least 0.994. These data showed excellent reproducibility. Inter-assay precision values at concentrations of 10, 40, and 80 ng/mL were 10.9%, 5.7%, and 2.6%, respectively, for oxycodone, and 9.6%, 6.3%, and 0.3%, for noroxycodone. Inter-assay accuracy at concentrations of 10, 40, and 80 ng/mL were –5.3%, 3.4%, and –0.4% for oxycodone, and –2.4%, 0.3%, and –1.3% for noroxycodone, respectively. The lower limit of quantification, defined as the lowest standard solution, was 0.5 ng/mL for oxycodone and noroxycodone.
Genotyping
Genomic deoxyribonucleic acid (DNA) was prepared from clots using the QIAamp® DNA Blood Mini Kit (Qiagen, CA, USA). Purifed genomic DNA was stored at –30°C prior to genotyping. Genotyping was carried out for the following single-nucleotide polymorphisms (SNPs): cytochrome P450 (CYP) 3A4*1G (C20230T: rs2242480), CYP3A5*3 (T6986C: rs776746) and CYP2D6*10 (C100T: rs1065852) (major oxycodone-metabolizing enzymes); P-glycoprotein [ATP-binding cassette, sub-family B member 1 (ABCB1)] C1236T (rs1128503), G2677T/A (rs2032582), C3435T (rs1045642) (major contributor to oxycodone excretion function); opioid receptor μ1 (OPRM1) A118G (rs1799971) (the primary site of action for oxycodone). Allelic variations were determined using the TaqMan® allelic discrimination assay (TaqMan® 5’-exonuclease allelic discrimination assay; Applied Biosystems, CA, USA). Genomic DNA (1 μL) was added to TaqMan® probe (Applied Biosystems), 2 × TaqMan® Universal PCR Master Mix (Applied Biosystems) and Distilled, Deionized, Sterile Water (Wako Pure Chemical Industries, Ltd., Japan). Gene fragments were amplified by polymerase chain reaction (PCR) using the StepOne® Real- Time PCR System. Amplification conditions consisted of an initial denaturation step for 10 min at 95°C, followed by 50 cycles of denaturation at 92°C for 15 sec and annealing and extension at 58°C for 1 min.
Statistical analysis
Association analysis between patient demographic characteristics and plasma concentration of oxycodone (ng/mL)/total daily dose (mg/day)/body weight (kg) was performed using Pearson's correlation or Student's t test with Bonferroni correction. The effects of severity of cachexia and genetic polymorphisms on metabolic activity were evaluated by Student's t test with Bonferroni correction. Genotype and allele frequencies for each polymorphism in the respective genes and the significance of deviations from Hardy-Weinberg equilibrium were calculated using Haploview 4.2. For determining linkage disequilibrium among the polymorphisms, pairwise linkage disequilibrium coefficients were calculated by the χ2 test. The influence of each genetic polymorphism on analgesic and adverse effects of oxycodone was statistically analyzed using the Mann-Whitney U test or Student’s t-test. Values of p<0.05 were considered as statistically significant.
Ethics approval and consent to participate
This study was carried out in accordance with the precepts of the Helsinki Declaration, and was approved by the ethics committee of Nagoya University Graduate School of Medicine (No. 947) and performed according to Good Clinical Practice Guidelines. The written informed consent documents were obtained from all subjects.
Results
Patient characteristics and oxycodone-induced adverse effects
Characteristics of fifty Japanese patients are shown in Table 1.
| Patient demographic characteristics | ||
|---|---|---|
| Characteristics | Median (interquartile range) (n=50) | Association with C/D ratio of oxycodone (p-value) |
| Sex: male | 30 [0.014 (0.01-0.02)] | 1 |
| Female | 20 [0.010 (0.00-0.03)] | |
| Age (years) | 63.5 (53.3-68.0) | 0.92 |
| Height (cm) | 160.3 (154.0-168.1) | 1 |
| Body weight (kg) | 78.2 (64-92.4) | 0.46 |
| Body mass index (kg/m2) | 20.6 (18.0-23.8) | 0.92 |
| Body surface area (m2) | 1.58 (1.39-1.67) | 1 |
| Blood urea nitrogen (mg/dL) | 14.5 (12.0-18.0) | 1 |
| Creatinine (mg/dL) | 0.74 (0.58-0.88) | 1 |
| Estimated glomerular filtration ratio (mL/min/1.73 m2) | 78.2 (63.5-93.3) | 1 |
| Aspartate transaminase (IU/L) | 22.0 (18.8-31.0) | 1 |
| Alanine Aminotransferase (IU/L) | 20.0 (13.8-33.0) | 1 |
| γ-Glutamyltranspeptidase (IU/L) | 34.0 (21.8-61.5) | 1 |
| Alkaline phosphatase (IU/L) | 312.0 (216.8-357.5) | 1 |
| Bilirubin (mg/dL) | 0.5 (0.4-0.6) | 1 |
| Albumin (g/dL) | 3.8 (3.5-4.1) | 1 |
| Ca (mEq/L) | 4.6 (4.4-4.7) | 1 |
| Hemoglobin (g/dL) | 11.2 (10.5-12.2) | 0.69 |
| Neutrophic leukocyte (× 103/μL) | 2.5 (1.6-3.6) | 1 |
| C-reactive protein (mg/dL) | 0.3 (0.1-0.8) | 1 |
| Child-Pugh score | 5 (5-6) | 1 |
| Performance status | 1 (1-1) | 1 |
| Pain intensity (NRS) | 1 (0-3) | 1 |
| Interval between the first and second interviews (day) | 14 (7.0-18.5) | 0.69 |
| Tumor type | Number of patients (n=50) | |
| Pancreatic cancer | 15 | |
| Colorectal cancer | 10 | |
| Breast cancer | 5 | |
| Lung cancer | 3 | |
| Hematological cancer | 3 | |
| Prostate cancer | 2 | |
| Uterine cancer | 2 | |
| Thyroid cancer | 1 | |
| Anal cancer | 1 | |
| Renal cancer | 1 | |
| Adrenal cancer | 1 | |
| Esophageal cancer | 1 | |
| Gastrointestinal stromal tumor | 1 | |
| Pheochromocytoma | 1 | |
| Sarcoma | 1 | |
| Orbital tumor | 1 | |
| unknown | 1 | |
Table 1: Patient demographic characteristics.
The most frequent type of tumor was pancreatic cancer (30%). The median oxycodone maintenance dose was 20 mg (with a range of 10-90 mg) and the median rescue dose was 2.5 mg (with a range of 2.5-15 mg) (Table 2a).
Adverse effects induced by oxycodone are shown in Table 2b. The number of patients experiencing nausea was 10 (grade 1: n=9, grade 2: n=1);constipation was 41 (grade 1: n=12, grade 2: n=29), and drowsiness was 23 (grade 1: n=22, grade 2: n=1).
| (a) Characteristics | Number of patients | Median (interquartile range) |
| Maintenance dose (mg/day) | 50 | 20 (10-90) |
| Rescue dose (mg) | ||
| No user | 27 | 0 |
| Use | 23 | 2.5 (2.5-15) |
| (b) Adverse reactions | Grade | Number of patients |
| Nausea | 0 - 1 - 2 - 3 | 40 - 09 - 01 - 0 |
| Constipation | 0 - 1 - 2 - 3 | 09 - 12 - 29 - 0 |
| Drowsiness | 0 - 1 - 2 - 3 | 27 - 22 - 01 - 0 |
Table 2: Oxycodone dosage (a) and the number of patients experienced oxycodone-induced adverse effects (b)
Allele frequency of CYP3A4, CYP3A5, CYP2D6, OPRM1, and ABCB1
The genotype and allele frequencies of polymorphisms are shown in Table 3. The minor allele frequencies were as follows: 0.22 for CYP3A4*1G, 0.23 for CYP3A5*3, 0.29 for CYP2D6*10, 0.48 (A) for A118G (OPRM1), 0.45 for C1236T (ABCB1), and 0.35 for C3435T (ABCB1). The allele frequency of G2677T/A (ABCB1) was 0.43 (G), 0.39 (T), 0.18 (A). Seven SNPs were identified in all patients (n=50), and SNPs in both the general population and patients were in Hardy-Weinberg equilibrium.
| Gene symbol | location | RefSNP ID | Exchange (position: GRCh38.p12) | Genotype (%) | Allele (%) | HWE |
|---|---|---|---|---|---|---|
| CYP3A4 | Intron 10 | rs2242480 | C20230T (chr7:99763843) | CC:31, CT:16, T:3 | C:78/T:22 | 0.85 |
| CYP3A5 | Intron 3 | rs776746 | T6986C (chr7:99672916) | TT:3, CT:17, CC:30 | T:23/C:77 | 1 |
| CYP2D6 | Exon 1 | rs1065852 | C100T (chr22:42130692) | CC:26, CT:19, TT:5 | C:71/T:29 | 0.77 |
| OPRM1 | Exon 1 | rs1799971 | A118G (chr6:15403966) | AA:11, AG: 26, GG:13 | A:48/G:52 | 1 |
| ABCB1 | Exon 1 | rs1128503 | C1236T (chr7:87550285) | CC:9, CT:27, TT:14 | C:45/T:55 | 0.78 |
| ABCB1 | Exon 21 | rs2032582 | G2677T/A (chr7:87531302) | GG:8, GT:10, TT:2,GA:10, AT:17, AA:5 | G:43/T:22/A:35 | - |
| ABCB1 | Exon 26 | rs1045642 | C3435T (chr7:87509329) | CC:22, CT:21, TT:7 | C:65/T:35 | 0.75 |
Table 3: Target gene polymorphisms and their allele frequencies.
Association between oxycodone dosage and polymorphisms in CYP3A4, CYP3A5, or CYP2D6 (genes involved in oxycodone pharmacokinetics/metabolism)
The influence of polymorphisms on oxycodone dose is shown in Figure 1A. We found several polymorphisms that had a tendency to increase the average total daily dose of oxycodone, as follows: CYP3A4 *1G/*1G (2.3 times increased, p=0.07); CYP3A5 *1/*1 (2.3 times increased, p=0.07); CYP2D6 100CC+CT (2 times increased, p=0.09). These tendencies were not observed when the data corrected with body weight (Figure 1B).
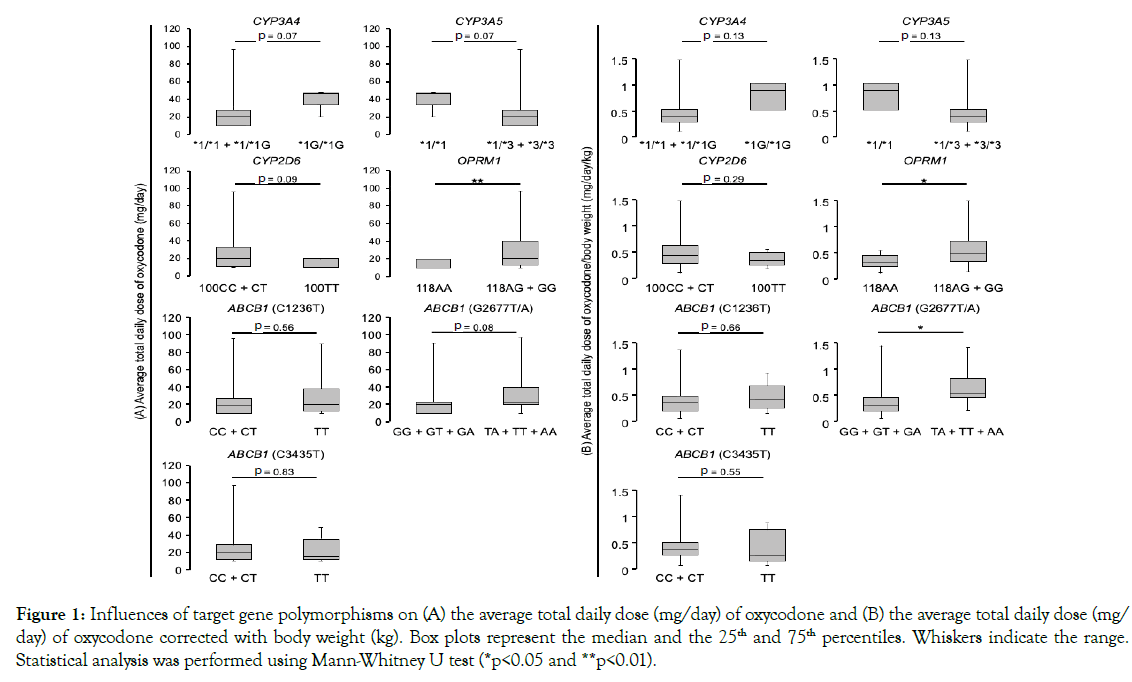
Figure 1. Influences of target gene polymorphisms on (A) the average total daily dose (mg/day) of oxycodone and (B) the average total daily dose (mg/day) of oxycodone corrected with body weight (kg). Box plots represent the median and the 25th and 75th percentiles. Whiskers indicate the range. Statistical analysis was performed using Mann-Whitney U test (*p<0.05 and **p<0.01).
The number of rescues in patients with CYP3A4 *1G/*1G and CYP3A5 *1/*1 was significantly higher than those with CYP3A4 *1/*1+*1/*1G and CYP3A5 *1/*3+*3/*3 (p<0.05) (Figure 2), but such differences were not observed in patients with CYP2D6 (p=0.17) (Figure 2). There was no significant association between pain relief ⊿NRS: CYP3A4 p=0.44, CYP3A5 p=0.44, and CYP2D6 p=0.29) (Figure S1) or degree of oxycodone-induced adverse effects (constipation: CYP3A4 p=0.61, CYP3A5 p=0.61, and CYP2D6 p=0.27; nausea: CYP3A4 p=0.58, CYP3A5 p=0.58, and CYP2D6 p=0.24; drowsiness: CYP3A4 and CYP3A5 p=0.50, CYP2D6 p=0.75) with any polymorphism (Figures S2-S4).
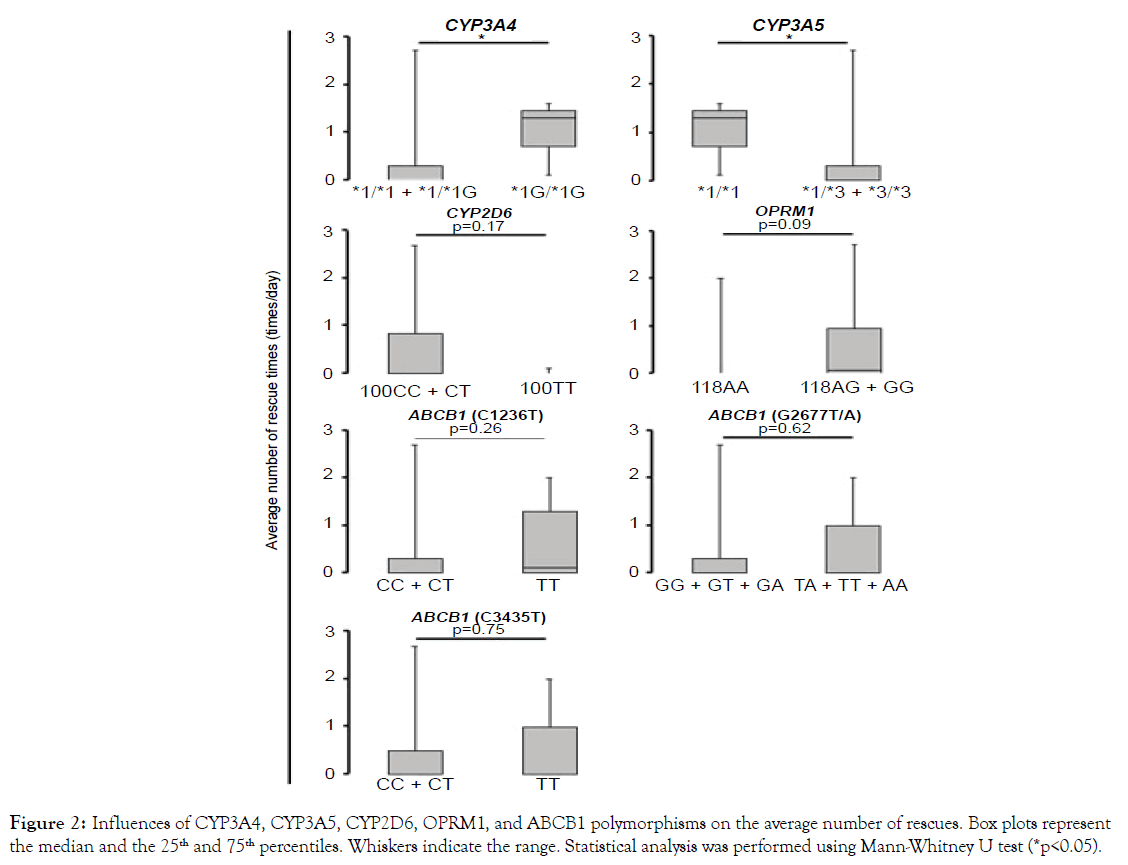
Figure 2. Influences of CYP3A4, CYP3A5, CYP2D6, OPRM1, and ABCB1 polymorphisms on the average number of rescues. Box plots represent the median and the 25th and 75th percentiles. Whiskers indicate the range. Statistical analysis was performed using Mann-Whitney U test (*p<0.05).
Association between oxycodone dosage and the OPRM1 (A118G) polymorphism (affecting oxycodone pharmacodynamics)
The average total daily dose of oxycodone received in patients with the OPRM1 118AG+GG alleles were increased (2 times) compared to those with the OPRM1 118AA allele (p<0.01) (Figure 1A) even when the data corrected with body weight (p<0.05) (Figure 1B). The number of rescues in patients with the OPRM1 118AG+GG alleles were likely to be increased, compared to those with the OPRM1 118AA allele (p=0.09) (Figure 2). No significant differences were observed in report of pain relief ⊿NRS: p=0.50) (Figure S1) or degree of adverse effects (constipation: p=0.96; nausea: p=0.30; drowsiness: p=0.44) in patients with OPRM1 polymorphisms (Figures S2-S4).
Association between oxycodone dosage and ABCB1 (C1236T, G2677T/A, C3435T) polymorphisms (genes involved in oxycodone pharmacokinetics/excretion)
Patients with the ABCB1 2677TA+TT+AA alleles had a tendency to receive a higher average total daily dose of oxycodone (1.1 times increased) compared to those with the ABCB1 2677 G allele (p=0.08) (Figure 1A). When the data corrected with body weight, the average total daily dose of oxycodone received in patients with the ABCB1 2677TA+TT+AA alleles were increased compared to those with the ABCB1 2677 G allele (p<0.05) (Figure 1B). Patients with the ABCB1 2677 G allele tended to exhibit greater pain relief than those with the ABCB1 2677TA+TT+AA alleles (2677TA+TT+AA: ⊿NRS=0.07, 2677GG+GT+GA: ⊿NRS=–0.58, p=0.08) (Figure S1).
The number of rescues (p=0.62) and the degree of adverse effects (constipation: p=0.29; nausea: p=0.12; drowsiness: p=0.98) did not statistically correlate with the ABCB1 G2677T/A polymorphism (Figures 2 and S2-4). Other ABCB1 polymorphisms (C1236T and C3435T) were not also associated with any analgesic responses (total daily dose: p=0.56 and p=0.83, respectively; the number of rescue: p=0.26 and p=0.75, respectively; ⊿NRS: p=0.58 and p=0.84, respectively) (Figures 1, 2, and S1) or adverse responses (constipation: p=0.41 and p=0.49, respectively; nausea: p=0.91 and p=0.67, respectively; drowsiness: p=0.79 and p=0.82, respectively) to oxycodone (Figures S2-4).
Association between genotype and plasma concentration of oxycodone
To evaluate the plasma concentration of oxycodone in 50 patients, we calculated as follows: ratio=plasma concentration of oxycodone (ng/mL)/average total daily dose of oxycodone (mg/day) and also the data corrected with body weight (kg). As shown in Table 4, the ratios in patients with CYP3A4 *1G/*1G and CYP3A5 *1/*1 were significantly higher than those with CYP3A4 *1/*1+*1/*1G (P<0.05) and CYP3A5 *1/*3+*3/*3 (p<0.05). When the data corrected with body weight, CYP3A4 *1G/*1G and CYP3A5 *1/*1 had tendencies to reach higher concentration than those with CYP3A4 *1/*1+*1/*1G (P=0.053) and CYP3A5 *1/*3+*3/*3 (p=0.053). In contrast, other polymorphisms failed to affect the plasma concentration of oxycodone.
| Plasma concentration of oxycodone/total daily dose | p- value | Plasma concentration of oxycodone/ total daily dose/body weight |
p-value | ||
|---|---|---|---|---|---|
| CYP3A4 | *1/*1+*1/*1G | 0.74 (0.43-1.16) | 0.007 | 0.011 (0.01-0.02) | 0.053 |
| *1G/*1G | 2.01 (1.58-2.62) | 0.037 (0.02-0.05) | |||
| CYP3A5 | *1/*1 | 2.01 (1.58-2.62) | 0.007 | 0.037 (0.02-0.05) | 0.053 |
| *1/*3+*3/*3 | 0.74 (0.43-1.16) | 0.011 (0.01-0.02) | |||
| CYP2D6 | 100CC+CT | 0.78 (0.43-1.17) | 0.42 | 0.012 (0.01-0.02) | 0.14 |
| 100TT | 1.16 (0.44-2.09) | 0.020 (0.01-0.06) | |||
| OPRM1 | 118AA | 0.82 (0.42-1.03) | 0.93 | 0.011 (0.01-0.03) | 0.82 |
| 118AG+GG | 0.76 (0.43-1.21) | 0.013 (0.01-0.03) | |||
| ABCB1 (C1236T) | 1236CC+CT | 0.76 (0.42-1.17) | 0.34 | 0.011 (0.01-0.02) | 0.47 |
| 1236TT | 0.90 (0.46-1.37) | 0.016 (0.01-0.03) | |||
| ABCB1 G2677T/A) | 2677GG+GT+GA | 0.78 (0.46-1.20) | 0.94 | 0.012 (0.01-0.02) | 0.98 |
| 2677TA+TT+AA | 0.79 (0.42-1.18) | 0.013 (0.01-0.025) | |||
| ABCB1 (C3435T) | 3435CC+CT | 0.78 (0.43-1.19) | 0.28 | 0.012 (0.01-0.02) | 0.66 |
| 3435TT | 0.85 (0.58-1.59) | 0.012 (0.01-0.04) |
Table 4: Influences of CYP3A4 and CYP3A5 polymorphisms on the plasma concentration of oxycodone/average total daily dose
To evaluate the differential effects of severity of cachexia and genetic polymorphisms on metabolic activity, we classified cachexia stage of patients, calculated the ratio of noroxycodone/oxycodone, and performed an association analysis. There was no statistically significant association between genetic polymorphisms and ratio of noroxycodone/oxycodone (Table S1).
Discussion
Sensitivity to physiological nociceptive and clinical pain differs considerably between individuals. Increasingly, part of this variability is recognized as an indication of genetic differences, which may mediate susceptibility to both the analgesic and adverse effects of treatments [21]. A high rate of interindividual difference in the activity of metabolizing enzymes, such as CYP3A and CYP2D6, due to genetic polymorphisms is well established. CYP3A and CYP2D6 are two major drug-metabolizing enzymes responsible for the oxidation of oxycodone. CYP3A4*1G (rs2242480), CYP3A5*3 (rs776746), and CYP2D6*10 (rs1065852) are highfrequency alleles and important enzymes in the metabolism of oxycodone, particularly in the Japanese population. Characterization of the effect of these particular alleles on oxycodone metabolism is clinically important, as it may help to predict individual responses to the drug [22-25]. In the present study, we found that patients with the CYP3A4 *1G/*1G or CYP3A5 *1/*1 genotypes had a tendency to receive a greater total daily dose of oxycodone and that the number of rescues was increased as compared to individuals with other genotypes. When we checked linkage disequilibrium (LD) profiles between rs2242480 (Chr:7, pos (hg38):99763843) and rs776746 (Chr:7, pos (hg38):99672916) using HaploReg v4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php), r2 and d' were 0.47 and 0.74, respectively, suggesting that these SNPs are independent relationship. However, since the carrier of CYP3A4 *1G/*1G and CYP3A5 *1/*1 were the same and only three subjects in the present results, caution is needed in the interpretation of our results. It has previously been reported that the CYP3A4*1G genetic polymorphism decreases CYP3A activity [6-8] and that the CYP3A5*3 genetic polymorphism results in almost the complete absence of CYP3A5 [9,10,26,27]. These studies reveal that patients with CYP3A4 *1G/*1G or CYP3A5 *3/*3 show increased oxycodone plasma concentrations and increased responsiveness to the drug, thereby decreasing the necessary dosage. However, patients with CYP3A4 *1G/*1G and CYP3A5 *1/*3 or *3/*3 genetic polymorphism did not exhibit high plasma concentration, and instead required decreased total daily dose of oxycodone and also decreased number of rescues. Therefore, there is still controversy concerning the impact of CYP3A4*1G and CYP3A5*3 genetic polymorphisms on oxycodone effectiveness. It is possible that some drugs, which act as substrates for CYP3A, could modulate the PK of oxycodone via drug-drug interactions [28,29]. There is also a possibility that the CYP3A5 polymorphism contributes to oxycodone PK rather than CYP3A4 [8]. CYP3A4 and CYP3A5 regions reveal that CYP3A4*1G is most often linked to CYP3A5*1, but rarely to CYP3A5*3 [8,30]. These studies suggest that environmental factors in addition to genetic factors are also associated with an individual’s response to oxycodone. We additionally examined concomitant agents (Tables S2 and S3) and other patients' conditions (Tables 1 and S1), and evaluated the association between plasma concentration of oxycodone (ng/mL)/total daily dose (mg/day)/body weight (kg) and use of concomitant agents and patients' conditions using Pearson's correlation analysis or Student's t-test with Bonferroni correction. There was no significant association between them, which potency affects PK/PD of oxycodone. Although the patients with CYP3A4 *1G/*1G and CYP3A5 *1/*1 commonly had took dexamethasone, a CYP3A4 moderate inducer (Table S3), there was no significant association between use of dexamethasone and the ratio of noroxycodone/oxycodone. In addition, the ratio of noroxycodone/oxycodone was not significantly associated with cachexia stage and genetic polymorphism. Further studies will be needed to clarify the influence of drug-drug interactions, patients' conditions, and/or association between CYP3A4 and CYP3A5 genetic polymorphisms using large sample size cohort to emphasize the evidence. The CYP2D6*10 genetic polymorphism leads to an unstable enzyme with lower metabolic activity [11] and individuals homozygous for *10 are phenotypically considered intermediate metabolizers (IM) [10,31,32]. Since CYP2D6 100TT leads to lower CYP2D6 activity, patients with 100TT are associated with decreased oxycodone doses. We found that the total daily dose of oxycodone and the number of rescues in patients with 100TT had a tendency to be lower compared to patients with 100CC or 100CT. A relatively increased plasma concentration of oxycodone in patients with CYP2D6 100TT was consistent to previous studies [10,11,31,32]. Because increased plasma concentration in patients with CYP2D6 100TT presumably leads to decreased daily dose and number of rescues, it suggests that the result of higher plasma concentration of CYP2D6 100TT carriers is highly reliable. There is, however, a shortcoming associated with these findings related to the CYP2D6 polymorphism. In the present study, only three CYP2D6*10 genotypes (100CC and CT and TT) were investigated, despite the frequency of other polymorphisms that affect CYP2D6 activity with relatively high frequency in Japanese populations (e.g. CYP2D6*5) [33]. We will need to assess the association between other CYP2D6 genetic polymorphisms and response to oxycodone to address this issue further. Many opioids, including oxycodone, are substrates of ABCB1, a major factor affecting transport, uptake, and PK/PD [34]. C1236T, C3435T, and G2677T/A are the most commonly studied SNPs in the ABCB1. Fujita et al. [12] and Takashina et al. [35] have shown that the TT genotype at 1236 in ABCB1 is associated with lower frequency of adverse effects and lower number of rescues. It seems that the C1236T polymorphism is associated with functional impairment of P-glycoprotein and is responsible for proper excretion. In contrast, the TT genotype of the C3435T polymorphism is associated with lowered ABCB1 expression [13,36]. Coulbault et al. [37] have shown an association between the G allele of G2677T/A and a reduced level of antiemetics use in postoperative pain treatment with morphine. These studies suggest that response to opioids and inclination for adverse effects are associated with ABCB1 genetic polymorphisms. In the present study, however, patients with the 2677 G allele had a tendency to receive a lower daily dose of oxycodone and to show greater pain relief as compared to those with other alleles. We also did not find an association between response to oxycodone and other ABCB1 polymorphisms (C1236 and C3435T). Although the clinical roles of ABCB1 polymorphisms are not obvious based on these discrepancies [13,35,38], some researchers have assumed that carriers of the combined genotype of the ABCB1 SNPs have enhanced differences in excretion functions [12,38]. To date, the functional effects of C1236T, G2677T/A, and C3435T in the ABCB1 gene on PK, efficacy and adverse effects of oxycodone remain controversial. Further studies are necessary to elucidate the roles of these polymorphisms on the functions of ABCB1. The opioid receptor μ1, encoded by OPRM1, is the primary site of action for the most commonly used opioids [12,13,35,38]. The OPRM1 is known to be the major action site specifically in oxycodone PD and the most prevalent SNP in the OPRM1 is a nucleotide substitution at position 118 (A118G). The G allele is more often found in poor responders to opioids like morphine or oxycodone and these patients require higher doses of opioids to relieve pain [12-18]. Our data indicate that patients with the OPRM1 118AG or GG received significantly greater daily doses of oxycodone and had a tendency to increase the number of rescues, compared to those with the 118AA genotype. These results suggest that the response of the opioid receptor μ1 in patients with the G allele is lower than in those with the AA genotype, which is consistent with several studies [12-18]. Thus, we demonstrate here that genetic polymorphisms of the OPRM1 are clinically associated with opioid effects. The present study has several limitations. While previous studies have investigated the association of genetic polymorphisms (such as combination of ABCB1 genetic polymorphisms [12,13] or linkage between CYP3A4 and CYP3A5 [8,22,30,39]), we focused on single genetic polymorphisms in the present study. Therefore, we consider that combined genetic polymorphisms could more accurately evaluate genetic influence on response to oxycodone and adverse effects associated with treatment compared to analysis of single genetic polymorphisms. Another issue is the influence of combined drug treatments on oxycodone PK. Drug-drug, drug-disease, and drug-food interactions related to functional induction and inhibition of CYP or P-glycoprotein may influence oxycodone PK. Accordingly, the present study should be conducted in a more extensive population and with more analysis (e.g. potential confounders or interactions).
Conclusions
These findings illustrate the importance of characterizing the effects of CYP3A, CYP2D6, ABCB1, and OPRM1 polymorphisms on the metabolism and effect of clinically important drugs such as oxycodone. Further studies that clarify the association between genetic polymorphisms related to the PK/PD of oxycodone and clinical patient’s phenotypes (symptoms, treatment response, adverse effects, drug-drug interactions, and so on) are needed.
Funding
This study was supported in part by Meijo University Research Institute Grant, and by Grants-in-Aid for Scientific Research C (24590219, 26460240, 16K08421, 17K10325) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
Acknowledgments
We would like to thank Dr. Akihiro Mouri and Dr. Aya Torii for helpful advice and Ms. Shoko Tsuchiya, Ms. Chihiro Murosaki, Ms. Haruka Kurishita, and Ms. Miho Asai for technical assistance.
REFERENCES
- Caraceni A, Hanks G, Kaasa S, Bennett MI, Brunelli C. Use of opioid analgesics in the treatment of cancer pain: evidence-based recommendations from the EAPC. Lancet Oncol. 2012;13:58-68.
- Samer CF, Daali Y, Wagner M, Hopfgartner G, Eap CB. Genetic polymorphisms and drug interactions modulating CYP2D6 and CYP3A activities have a major effect on oxycodone analgesic efficacy and safety. Br J Pharmacol. 2010;160:919-930.
- Lalovic B, Phillips B, Risler LL, Howald W, Shen DD. Quantitative contribution of CYP2D6 and CYP3A to oxycodone metabolism in human liver and intestinal microsomes. Drug Metab Dispos 2004;32:447-454.
- Cone EJ, Darwin WD, Buchwald WF, Gorodetzky CW. Oxymorphone metabolism and urinary excretion in human, rat, guinea pig, rabbit, and dog. Drug Metab Dispos. 1983;11:446-450.
- Lalovic B, Kharasch E, Hoffer C, Risler L, Liu-Chen LY. Pharmacokinetics and pharmacodynamics of oral oxycodone in healthy human subjects:role of circulating active metabolites. Clin Pharmacol Ther. 2006;79:461-479.
- Zhang W, Chang YZ, Kan QC, Zhang LR, Li ZS. CYP3A4*1G genetic polymorphism influences CYP3A activity and response to fentanyl in Chinese gynecologic patients. Eur J Clin Pharmacol. 2010;66:61-66.
- Gao Y, Zhang LR, Fu Q. CYP3A4*1G polymorphism is associated with lipid-lowering efficacy of atorvastatin but not of simvastatin. Eur J Clin Pharmacol. 2008;64:877-882.
- Sai K, Saito Y, Fukushima-Uesaka H, Kurose K, Kaniwa N. Impact of CYP3A4 haplotypes on irinotecan pharmacokinetics in Japanese cancer patients. Cancer Chemother Pharmacol. 2008;62:529-537.
- Tsuchiya N, Satoh S, Tada H, Li Z, Ohyama C. Influence of CYP3A5 and MDR1 (ABCB1) polymorphisms on the pharmacokinetics of tacrolimus in renal transplant recipients. Transplantation. 2004;78:1182-1187.
- Naito T, Takashina Y, Yamamoto K, Tashiro M, Ohnishi K. CYP3A5*3 affects plasma disposition of noroxycodone and dose escalation in cancer patients receiving oxycodone. J Clin Pharmacol. 2011;51:1529-1538.
- Ishiguro A, Kubota T, Ishikawa H, Iga T. Metabolic activity of dextromethorphan O-demethylation in healthy Japanese volunteers carrying duplicated CYP2D6 genes:duplicated allele of CYP2D6*10 does not increase CYP2D6 metabolic activity. Clin Chim Acta. 2004;344:201-204.
- Fujita K, Ando Y, Yamamoto W, Miya T, Endo H. Association of UGT2B7 and ABCB1 genotypes with morphine-induced adverse drug reactions in Japanese patients with cancer. Cancer Chemother Pharmacol. 2010;65:251-258.
- Campa D, Gioia A, Tomei A, Poli P, Barale R. Association of ABCB1/MDR1 and OPRM1 gene polymorphisms with morphine pain relief. Clin Pharmacol Ther. 2008;83:559-566.
- Chou WY, Yang LC, Lu HF, Ko JY, Wang CH. Association of mu-opioid receptor gene polymorphism (A118G) with variations in morphine consumption for analgesia after total knee arthroplasty. Acta Anaesthesiol Scand. 2006;50:787-792.
- Hayashida M, Nagashima M, Satoh Y, Katoh R, Tagami M. Analgesic requirements after major abdominal surgery are associated with OPRM1 gene polymorphism genotype and haplotype. Pharmacogenomics. 2008;9:1605-1616.
- Klepstad P, Rakvag TT, Kaasa S, Holthe M, Dale O. The 118 A>G polymorphism in the human mu-opioid receptor gene may increase morphine requirements in patients with pain caused by malignant disease. Acta Anaesthesiol Scand. 2004;48:1232-1239.
- Lotsch J, Skarke C, Grosch S, Darimont J, Schmidt H. The polymorphism A118G of the human mu-opioid receptor gene decreases the pupil constrictory effect of morphine-6-glucuronide but not that of morphine. Pharmacogenetics. 2002;12:3-9.
- Sia AT, Lim Y, Lim EC, Goh RW, Law HY. A118G single nucleotide polymorphism of human mu-opioid receptor gene influences pain perception and patient-controlled intravenous morphine consumption after intrathecal morphine for postcesarean analgesia. Anesthesiology. 2008;109:520-526.
- Hanks GW, Reid C. Contribution to variability in response to opioids. Support Care Cancer. 2005;13:145-152.
- Kasai S, Hayashida M, Sora I, Ikeda K. Candidate gene polymorphisms predicting individual sensitivity to opioids. Naunyn Schmiedebergs Arch Pharmacol. 2008;377:269-281.
- Kehlet H, Jensen TS, Woolf CJ. Persistent postsurgical pain:risk factors and prevention. Lancet. 2006;367:1618-1625.
- Fukushima-Uesaka H, Saito Y, Watanabe H, Shiseki K, Saeki M. Haplotypes of CYP3A4 and their close linkage with CYP3A5 haplotypes in a Japanese population. Hum Mutat. 2004;23:100.
- Nishida Y, Fukuda T, Yamamoto I, Azuma J. CYP2D6 genotypes in a Japanese population:low frequencies of CYP2D6 gene duplication but high frequency of CYP2D6*10. Pharmacogenetics. 2000;10:567-570.
- Daly AK. Significance of the minor cytochrome P450 3A isoforms. Clin Pharmacokinet. 2006;45:13-31.
- Hiratsuka M, Takekuma Y, Endo N, Narahara K, Hamdy SI. Allele and genotype frequencies of CYP2B6 and CYP3A5 in the Japanese population. Eur J Clin Pharmacol. 2002;58:417-421.
- Namazi S, Kojuri J, Khalili A, Azarpira N. The impact of genetic polymorphisms of P2Y12, CYP3A5 and CYP2C19 on clopidogrel response variability in Iranian patients. Biochem Pharmacol. 2012;83:903-908.
- Satoh S, Kagaya H, Saito M, Inoue T, Miura M. Lack of tacrolimus circadian pharmacokinetics and CYP3A5 pharmacogenetics in the early and maintenance stages in Japanese renal transplant recipients. Br J Clin Pharmacol. 2008;66:207-214.
- Nieminen TH, Hagelberg NM, Saari TI, Pertovaara A, Neuvonen M. Rifampin greatly reduces the plasma concentrations of intravenous and oral oxycodone. Anesthesiology. 2009 110:1371-1378.
- Saari TI, Gronlund J, Hagelberg NM, Neuvonen M, Laine K. Effects of itraconazole on the pharmacokinetics and pharmacodynamics of intravenously and orally administered oxycodone. Eur J Clin Pharmacol. 2010;66:387-397.
- de Forni M, Bugat R, Chabot GG, Culine S, Extra JM. Phase I and pharmacokinetic study of the camptothecin derivative irinotecan, administered on a weekly schedule in cancer patients. Cancer Res. 1994;54:4347-4354.
- Perwitasari DA, Wessels JA, van der Straaten RJ, Baak-Pablo RF, Mustofa M. Association of ABCB1, 5-HT3B receptor and CYP2D6 genetic polymorphisms with ondansetron and metoclopramide antiemetic response in Indonesian cancer patients treated with highly emetogenic chemotherapy. Jpn J Clin Oncol. 2011;41:1168-1176.
- Zanger UM, Raimundo S, Eichelbaum M. Cytochrome P450 2D6:overview and update on pharmacology, genetics, biochemistry. Naunyn Schmiedebergs Arch Pharmacol. 2004;369:23-37.
- Iwashima K, Yasui-Furukori N, Kaneda A, Saito M, Nakagami T. No association between CYP2D6 polymorphisms and personality trait in Japanese. Br J Clin Pharmacol. 2007;64:96-99.
- Hassan HE, Myers AL, Lee IJ, Coop A, Eddington ND. Oxycodone induces overexpression of P-glycoprotein (ABCB1) and affects paclitaxel's tissue distribution in Sprague Dawley rats. J Pharm Sci. 2007;96:2494-2506.
- Takashina Y, Naito T, Mino Y, Yagi T, Ohnishi K. Impact of CYP3A5 and ABCB1 gene polymorphisms on fentanyl pharmacokinetics and clinical responses in cancer patients undergoing conversion to a transdermal system. Drug Metab Pharmacokinet. 2012;27:414-421.
- Meineke I, Freudenthaler S, Hofmann U, Schaeffeler E, Mikus G. Pharmacokinetic modelling of morphine, morphine-3-glucuronide and morphine-6-glucuronide in plasma and cerebrospinal fluid of neurosurgical patients after short-term infusion of morphine. Br J Clin Pharmacol. 2002;54:592-603.
- Coulbault L, Beaussier M, Verstuyft C, Weickmans H, Dubert L. Environmental and genetic factors associated with morphine response in the postoperative period. Clin Pharmacol Ther. 2006;79:316-324.
- Zwisler ST, Enggaard TP, Noehr-Jensen L, Mikkelsen S, Verstuyft C. The antinociceptive effect and adverse drug reactions of oxycodone in human experimental pain in relation to genetic variations in the OPRM1 and ABCB1 genes. Fundam Clin Pharmacol. 2010;24:517-524.
- Miura M, Satoh S, Kagaya H, Saito M, Numakura K. Impact of the CYP3A4*1G polymorphism and its combination with CYP3A5 genotypes on tacrolimus pharmacokinetics in renal transplant patients. Pharmacogenomics. 2011;12:977-984.
Citation: Yoshimi A, Yoshijima Y, Miyazaki M, Kato H, Kato YK, Yamada K, et al. (2019). Association Study between Genes Related to Pharmacokinetics and Pharmacodynamics of Oxycodone and Response to Drug Treatment: A Genetic Cohort Study. J Pharmacovigil 7:275
Copyright: © 2019 Yoshimi A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Competing interests: The authors have declared that no competing interests exist.
Sources of funding : This study was supported in part by Meijo University Research Institute Grant, and by Grants-in-Aid for Scientific Research C (24590219, 26460240, 16K08421, 17K10325) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.