Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- CiteFactor
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Review - (2021) Volume 12, Issue 1
A Critical Review on Defense Mechanisms of Plants against Bacterial Pathogens: From Morphological to Molecular Levels
Tibebu Belete*Received: 07-Jan-2021 Published: 28-Jan-2021, DOI: 10.35248/2157-7471.21.12.534
Abstract
Better understanding of plant defense mechanism is crucial for improving crop health and yield. Plant defense against bacterial pathogens results from a complex combination of structural plant characteristics and induced biochemical reactions. In addition to the constitutive defense, plants may perceive directly or indirectly the presence of a bacterium and subsequently induce plant defense responses. These inducible biochemical reactions tend to create protective physiological conditions to limit bacterial growth and invasion in the host tissues. The inducible plant defense starts when a particular bacterial molecule or its structural feature is recognized by trans-membrane protein recognition receptors (PRRs) on plant cell surface. The recognition is based on conserved features of molecules of bacterial origin, namely pathogen associated molecular patterns (PAMPs). This induces PAMP-triggered immunity (PTI) and the expression of defense genes, what prevents pathogenesis. However, some pathogens may release effector molecules and surpass PTI what leads to effector-triggered susceptibility (ETS). Subsequently, plants possess resistance (R) proteins usually containing nucleotide-binding (NB) and leucine-rich repeat (LRR) domains which trigger signaling cascade by recognizing specific effectors. This leads to the activation of downstream genes in order to create a robust and fast defense response preventing the spread of bacteria. Generally, these actions against invading bacterial pathogen are controlled directly or indirectly by genetic materials (gene) of the host plants. Therefore, the objective of this review is to discuss and summarize how the receptors are thought to activate defenses, how bacterial pathogens surpass this basal defense system and how plants have evolved a second defense layer, with an emphasis on the future research priorities.
Keywords
Pathogenic bacteria; Defense mechanisms; PAMP; T3SS; PTI; EF-Tu; PRR
Introduction
Plants suffer from bacterial diseases caused by many bacterial pathogens from both the Proteobacteria and Actinobacteria. Some of the most common and most studied plant pathogenic genera of bacteria include Agrobacterium, Erwinia, Pseudomonas, Xanthomonas, Ralstonia, Clavibacter, Streptomyces and Xylella fastidiosa [1]. Of these, members of the genera Pseudomonas and Xanthomonas are causative agents of almost all bacterial spots and blights of leaves, stems, and fruits, while Agrobacterium is the main cause of grown gall on many woody plants [2]. Infected plants show a variety of symptoms, such as leaf spots and blights, soft rots, wilts, galls, specks, chlorosis (yellowing), cankers and cancers.
Bacteria can be sucked into a plant through natural plant openings such as trichomes, lenticels, stomata, hydathodes, nectarthodes or stigma. They can also enter through abrasions or wounds on leaves, stems or roots or through placement by specific feeding insects.
Artificially, bacteria are most commonly introduced into plants by wounding, by pressure-driven aerosols mimicking wind-driven rains, vacuum infiltration, or by seed immersion into inoculum [3]. Once inside the plant system, bacteria proliferate the apoplast, that is, the intercellular spaces or xylem vessels. Invasion of the apoplast results in parenchymatous and vascular or parenchymatous-vascular diseases. Some Pseudomonas syringae strains make coronatine, a jasmonic acid mimic that suppresses salicylic acid mediated defense to biotrophic pathogens and induces stomatal opening, helping pathogenic bacteria gain access to the apoplast [4].
Plants are continuously threatened by pathogen attack and, as such, they have evolved mechanisms to evade, escape and defend themselves against pathogens [5]. The moment pathogen inoculum come in contact with host surface, the plants due to hereditary characters have several naturally occurring physical and chemical barriers (pre-existing) resisting penetration, and if at all the penetration occurs, the host reacts by different means resulting in formation of physical and chemical barriers. Although lacking an immune system comparable to animals, plants have developed an innate immunity comprising several structural, chemical, and protein-based defenses designed to detect and stop invading organisms. Therefore higher plants protect themselves against bacterial infection or other biotic and abiotic factors in different ways. They defend themselves against such factors by physical strengthening of the cell wall through lignification, suberization, and producing various pathogenesis-related (PR) proteins [4].
Generally, plant defense starts when a particular pathogen molecule or its structural feature is recognized by trans-membrane protein recognition receptors (PRRs) on plant cell surface. The recognition is based on conserved features of molecules of bacterial or fungal origin, namely pathogen associated or microbial associated molecular patterns (PAMPs or MAMPs). This induces PAMP-triggered immunity (PTI) and the expression of defense genes, what prevents pathogenesis. However, some pathogens may release effector molecules and surpass PTI what leads to effectortriggered susceptibility (ETS). Subsequently, plants possess resistance (R) proteins usually containing nucleotide-binding (NB) and leucine-rich repeat (LRR) domains which trigger signaling cascade by recognizing specific effectors. This leads to the activation of downstream genes in order to create a robust and fast defense response preventing the spread of pathogens. The recognition of effector molecules by R proteins and the triggered defense response are known as effector-triggered immunity (ETI). The signals from effector recognition are transmitted to the nucleus where they promote defense gene expression. These may code for transcription factors to commence the transcription of downstream enzymes required for the production of defense related metabolites such as salicylic acid (SA) or pathogenesis related (PR) proteins. Such signal transduction pathways lead to hypersensitive response (HR) characterized by accumulation of SA, reactive oxygen species, and the synthesis of PR proteins. The HR results in programmed cell death to stop pathogen invasion [6].
Literature Review
In this review paper, we try to discuss and summarized how the receptors are thought to activate defenses, how bacterial pathogens surpass this basal defense system and how plants have evolved a second defense layer and giving directions for future research priorities.
Bacterial secretion systems and proteomic studies
In the last decades a lot of effort has been dedicated to elucidate the interaction between plants and bacteria, both beneficial and pathogenic bacteria rely on diverse secretion pathways in order to overcome plant defenses and to establish successful colonization of the host plant. Sive secretion systems (type I–VI) have been reported in bacteria, which are distinguished by their constituent proteins. The most widely studied and main secretion system used by pathogenic bacteria during infection is the type III secretion system (T3SS), which is involved in some of the most devastating pathogens in plants like Xanthomonas species and Pseudomonas pathovars [7,8]. It is a multi-protein complex related to bacterial flagellum and forms a pilus that injects effectors into the plant cell [9]. In other words, it enables bacteria to directly inject effector proteins or virulence factors into the plant cell and interrupt cellular processes. T3SS is essential for pathogenicity and is conserved amongst gram negative bacteria; however, the proteins exported by this system are more variable [10]. The best studied T3SS effectors are designated Avr proteins, which have been reported in several plant pathogens [9,11]. Other effectors have also been identified in different phytopathogenic bacterial species, including Xanthomonas outer protein (Xop) in Xanthomonas, hypersensitive reaction and pathogenicity (Hrp) outer protein (Hop) in Pseudomonas and Pseudomonas outer protein (Pop) (based on a previous genus designation) in Ralstonia [12].
Another important system for bacterial pathogenicity is the type II secretion system (T2SS), which is involved in the secretion of extracellular enzymes (pectinases, endo-glucanases and cellulases), toxins and virulence factors. Striking differences in the number and combinations of these enzymes in different pathogens are expected to be found. T2SS commonly found in the genus Erwinia commonly now a day Pectobacterium, is used for the secretion of cell wall-degrading enzymes causing soft-rot, while the type IV secretion system (T4SS) has a critical role in the pathogenesis of Agrobacterium tumefaciens and its capability to form galls on plants [7,9]. Severalbacterial proteins are transported through the T4SS to enable the efficient transfer andintegration of bacterial DNA. It is important to note that many pathogens rely on multiplemechanisms of protein secretion. For example, many Erwinia species require both a T2SSand a T3SS to cause disease, and several strains of Xanthomonas have T2SS, T3SS andT4SS [9] (Figure 1).
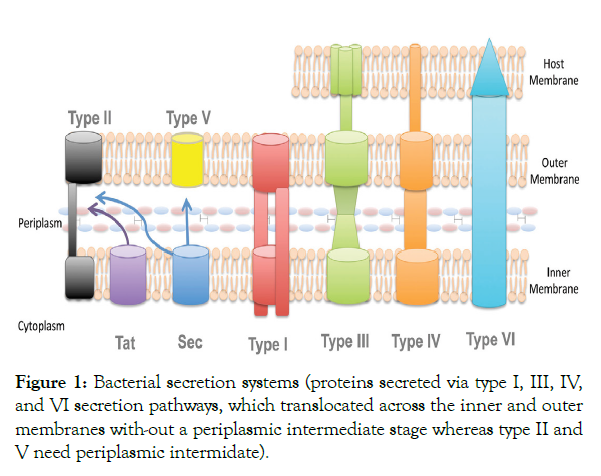
Figure 1: Bacterial secretion systems (proteins secreted via type I, III, IV, and VI secretion pathways, which translocated across the inner and outer membranes with-out a periplasmic intermediate stage whereas type II and V need periplasmic intermidate).
Most of the data currently available on pathogenicity mechanisms in bacteria have been obtained by genomic studies. Few studies as compared to genomic studies have employed the proteomic approach, which aims to identify the bacterial proteins putatively involved in pathogenicity. In recent years, proteomics has played a key role in identifying changes in protein levels in plant hosts upon infection by pathogenic organisms and in characterizing cellular and extracellular virulence and pathogenicity factors produced by pathogens [13]. Proteomics offers a constantly evolving set of novel techniques to study all aspects of protein structure and function. It aims to find out the identity and amount of each and every protein present in a cell and actual function mediating specific cellular processes. Mehta and Rosato [14] reported the analysis of X. axonopodis pv. citri cultivated in the presence of the host Citrus sinensis leaf extract, and identified differentially expressed proteins, including a sulfate-binding protein, by NH2 terminal sequencing. The authors suggested that the induction of this enzyme may have been caused by the amino acids or different sugars present in the leaf extract. Tahara et al. [15] also analyzed the expressed proteins of X. axonopodis pv. passiflorae during the interaction with the host Passiflorae edulis leaf extract, and identified an inorganic pyrophosphatase and an outer membrane protein up regulated in the presence of leaf extract, also by NH2 terminal sequencing. It was proposed that the outer membrane protein identified may have an important role in pathogenicity [15].
With regard to plant defense responses, direct evidence of the involvement of target proteins has also been provided by proteomic studies. Although few, the reports outlined below clearly show the importance of proteomic approaches, which can aid significantly in the understanding of plant bacterium interactions. A detail understanding of plant defense response using successful combination of proteomic techniques is needed for practical application to secure and stabilize yield of many crop plants [16]. Jones et al. [17] analyzed the proteomic and transcriptomic profiles of Arabidopsis thaliana leaves during early responses (1–6 hr. post-inoculation) to the challenge by P. syringae pv. tomato. They compared the proteomic changes in A. thaliana in response to the P. syringae pv. tomato highly virulent strain DC3000, which results in successful parasitism, a DC3000 hrp mutant, which induces basal resistance, and a trans-conjugant of DC3000 expressing avrRpm1, which triggers a gene-for-gene-based resistance. Two subsets of proteins, which consistently showed clear differences in abundance after various challenges and time intervals, were glutathione S-transferases (GSTs) and peroxiredoxins (Prxs). Both of these groups of antioxidant enzymes were considered to have probable significant roles in the regulation of redox conditions within infected tissue . These results were further related to changes in the expression profiles for the corresponding GST and Prx genes, identified by Affymetrix GeneChip analysis. In general, a good correlation was observed between changes obtained at the transcript and protein levels for the Prx family, but not for the GST family. Only for the PrxB protein was the decrease observed in the spot intensity following pathogen challenge clearly related to transcriptional suppression. These observations were used to highlight the complexity of comparative proteomics and transcriptomics, even when derived from the same inoculation system.
As a follow-up study, the same group [18], examined the global proteomic profile in three subcellular fractions (soluble protein, chloroplast and mitochondria enriched) of A. thaliana responding to the same three P. syringae pv. tomato DC3000 strains. This was the first report to associate post-translational events (1–6 hr. post-inoculation) occurring before significant transcriptional reprogramming. In total, 73 differential spots representing 52 unique proteins were successfully identified (Table 1), and were representative of two major functional groups: defense-related antioxidants and metabolic enzymes. The results showed that several chloroplast systems are modified during all aspects of the defense response. Components of the Calvin–Benson cycle are rapidly altered during basal defense, and some of these changes are reversed by type III effectors. Photosystem II has emerged as a target of resistance signaling. Mitochondrial porins appear to be modified early in basal defense, with specific alterations to other components in response to AvrRpm1. Finally, the interplay between redox status and glycolysis, with probable links to lipid signaling [through glyceraldehyde 3-phosphate dehydrogenase, some GSTs, lipase and NADH: quinone oxidoreductase (NQR)], may coordinate communication between organelles. Significant changes to photosystem II and to mitochondrial porins seem to occur early in basal defense. Rapid communication between organelles and the regulation of primary metabolism through redox-mediated signaling are supported by these results.
| No. of proteins identified | Plant species | Pathogens | Proteomic method used |
|---|---|---|---|
| 12 | Arabidopsis thaliana | P. syringe pv. Tomato DC3000 |
Nitration of proteins, 2D-PAGE, LC MS/MS |
| 52 | A. thaliana | P. syringae | 2D-PAGE, LC-MS/MS |
| 5 | Citrus sinensis | X. axonopodis pv. citri | 2D-PAGE, N-terminal sequencing |
| 20 | Glycine max | X. axonopodis pv. Glycine | 2D-PAGE |
| 47 | L. hirsutum | Clavibacter michiganensis sp. michiganensis | 2D-PAGE, LCQ-Deca ion trap MS |
| 20 | Oryza sativa | X. oryzae pv. oryzae | 2D-PAGE, MALDI TOF, Edman degradation |
| 7 | Olea europaea sub sp Europaea | Pseudomonas savastanoi pv. Savastanoi | 2D-PAGE, MALDI TOF |
| 4 | Passiflora edulis | X. axonopodis pv. Passiflorae | 2D- PAGE, Edman sequencing |
| 12 | Solanum lycopersicum | Ralstonia solanacearum | 2D- PAGE, n and cLC de novo |
| 41 | Saintpaulia ionantha | Dickeya dadantii (syn. E. chrysanthemi) | 2D-PAGE, MALDI-TOF MS |
Table 1: Some of the most important defense related identified number of proteins in proteomics [50].
Many studies were conducted between rice and bacterial association, some are pathogenic and cause severe damages to the crop, such as X. oryzae pv oryzae, Burkholderia glumae, Burkholderia kururiensis and P. fuscovaginae. Among them, Burkholderia kururiensis is very often isolated from rice and has been studied recently for its potential beneficial effects on the plant and the mechanisms of interaction [19]. To investigate the role of defense responsive proteins in the X. oryzae pv. oryzae interaction, Mahmood et al. [20] applied a proteomic approach. Cytosolic and membrane proteins were fractionated from the rice leaf blades 3 days post-inoculation with incompatible and compatible X. oryzae pv. oryzae races. From 366 proteins analyzed by 2DE, 20 were differentially expressed in response to bacterial inoculation (Table 1). Analyses clearly revealed that four defense related proteins [PR-5, probenazole-inducible protein (PBZ1), superoxide dismutase (SOD) and Prx] were induced for both compatible and incompatible X. oryzae pv. oryzae races, wherein PR-5 and PBZ1 were more rapid and showed higher induction in incompatible interactions and in the presence of jasmonic acid (JA). Studying the same rice X. oryzae pv. oryzae interaction, Chen et al. [21] analyzed proteins from rice plasma membrane to study the early defense responses involved in XA21-mediated resistance. XA21 is a rice receptor kinase, predicted to perceive the X. oryzae pv. oryzae signal at the cell surface, leading to the ‘gene-for-gene’ resistance response. They observed a total of 20 proteins differentially regulated by pathogen challenge at 12 and 24 hr. post-inoculation, and identified at least eight putative plasma membrane-associated and two non-plasma membrane-associated proteins with potential functions in rice defense (Table 1).
Proteins from the wild tomato species Lycopersicon hirsutum that are regulated in response to the causal agent of bacterial canker (Clavibacter michiganensis ssp. michiganensis) were identified by comparing two partially resistant lines and a susceptible control line in a time course (72 and 144 hr. post-inoculation) experiment [14]. Using 2DE and ESI-MS ⁄MS, 26 differentially regulated tomato proteins were identified, 12 of which were directly related to defense (Table 1).
Proteomic analysis was also used to detect the responses of the model legume Medicago truncatula to the pathogenic bacterium Pseudomonas aeruginosa in the presence of known bacterial quorum sensing signals, such as N-acyl homoserine lactone (AHL) [22]. The fast and reliable detection of bacterial AHL signals by plant hosts is essential to make appropriate responses to the pathogen. Therefore, M. truncatula is able to detect very low concentrations of AHL from P. aeruginosa, and responds in a global manner by significant changes in the accumulation of 154 proteins, 21 of which are related to defense and stress responses (Table 2).
| Protein | Studied organism | Pathogen |
|---|---|---|
| lutathione S-transferase | A. thaliana | P. syringae |
| Peroxiredoxin | A. thaliana | P. syringae |
| Peroxiredoxin, chloroplast | O. sativa | X. oryzae pv. oryzae |
| Glyceraldehyde 3-phosphate dehydrogenase |
O. sativa | X. oryzae pv. oryzae |
| Triosephosphate isomerase, cytosolic (EC 5.3.1.1) | O. sativa | X. oryzae pv. oryzae |
| Thaumatin-like protein X | O. sativa | X. oryzae pv. oryzae |
| Superoxide dismutase | O. sativa | X. oryzae pv. oryzae |
| Alcohol dehydrogenase 1 a | O. sativ | X. oryzae pv. oryzae |
| Quinone reductase | O. sativa | X. oryzae pv. oryzae |
| Prohibitin | O. sativa | X. oryzae pv. oryzae |
| Ascorbate peroxidase | O. sativa | X. oryzae pv. oryz |
| Remorin 1 | Lycopersicon hirsutum |
Clavibacter michiganensis ssp. michiganensis |
| Ascorbate peroxidase | L. hirsutum | C. michiganensis ssp. michiganensis |
| Glutathione S-transferase | L. hirsutum | C. michiganensis ssp. michiganensis |
| Pathogenesis-related 3 (endochitinase precursor) |
L. hirsutum | C. michiganensis ssp. michiganensis |
Table 2: Some examples of proteins expressed in plant–bacterial interactions and identified in plants using proteomic approaches [14,17,18,20,21].
Plant defense responses against bacterial pathogens
During the co-evolution of bacterial pathogens and their plant host, plants have developed intricate defense mechanisms against pathogen infections. They have evolved multiple defense strategies for combating invading pathogens such as: (1) Structural characteristics that act as physical barriers and inhibit the pathogen from gaining entrance and spreading through the plant and (2) biochemical reactions that take place in the cells and tissues of the plant and produce substances that are either toxic to the pathogen or create conditions that inhibit growth of the pathogen in the plant. The combination of structural characteristics and biochemical reactions employed in the defense of plants are different host-pathogen systems. However, whatever the kind of defense or resistance a host plant employs against a pathogen, it is ultimately controlled, directly or indirectly by the genetic material (genes) of the host plant and of the pathogen [23].
From non-host resistance to gene-for gene race specific resistance, defense responses in plants are layered and continuing responses and signal transduction pathways are overlapped. The first barrier of plants is natural passive defenses including the surface wax, stomata, cell wall and some pre-existing toxins and enzymes. Plants also develop the ability to express the induced defense responses through recognizing certain microbial molecules. This kind of molecules usually have repeated conserved features like lipopolysaccharides (LPS) and flagellin and are referred as pathogen associated molecular patterns (PAMP) or microbe-associated molecular patterns (MAMP) [24]. In other words, in a first layer of defense, conserved microbial molecules called PAMPs are recognized on the cell surface which then leads to the induction of a number of defense responses, including the generation of reactive oxygen species (ROS), the initiation of mitogen-activated protein kinase (MAPK) signaling, PR-gene expression, and callose depositions at the cell wall. Any invaders that overcome both barriers must still face the formidable task of overcoming the plant immune response. Plant immunity can be broken down into two components operating on different time scales. The basal defense system appears early in pathogen interaction, while the resistance (R) gene mediated defense operates on the time scale of hours [25].
Preformed structural and biochemical defenses
Preformed structural defense mechanisms: The preformed, first line of defense, pre-existing, non-inducible, passive or pre-invasive plant defense mechanisms are the innate basal first line immune defense gadgets indigenously constitutive in the plant even before colonization by the pathogen. The preformed structural defense mechanisms often found on the plant surface, are generally of the categories that present physical barriers to pathogen entry. The physical barriers include wax layers, rigid cell walls, stomata, epidermal layer, lenticels, cuticular lipids, cutin, etc [2]. These structures not only protect the plant from invasion, they also give the plant strength and rigidity and exist as integral component physiological structures throughout the lifespan of the plant [4].
Stomatal defense: Stomata are a small microscopic natural pores/openings in the epidermis of the areal part of plants that allow plants to exchange gases with the environment. At the same time they allow water loss by transpiration [9,26]. Historically, stomata have been considered as passive portal of entry for plant pathogenic bacteria. In other words, entry of bacteria into leaf tissues through natural openings has been generally perceived as a passive event, where bacteria lack active mechanisms for gaining entry, and plants similarly lack active mechanisms for preventing entry. However, recent studies suggest that stomata can play an active role in restricting bacterial invasion as part of the plant innate immune system [27]. Pathogens, such as the bacterium X. campestris pv. armoraciae, the oomycete Plasmopara viticola, and species of the fungus Puccinia are specialized to internalize into leaves only through stomata [28]. Earlier observations provided some clues that stomatal closure might diminish bacterial disease severity in a biologically relevant context. For instance, reduced number of lesions developed on dark or abscisic acid (ABA)-treated tomato plants after inoculation with X. campestris pv. vesicatoria. Interestingly, many bacterial disease outbreaks require high humidity, rain, or frost damage, which could promote stomatal opening and/or bypass stomatal defense by creating wounds as alternative entry sites [27].
Stomata respond to the presence of pathogenic bacteria and close rapidly [29] and this closure depends on the recognition of PAMPs (Figures 2 and 3) by cell surface pattern recognition receptors (PRRs). For example when P. syringae pv. tomat (Pst) gains access to the apoplast via stomata, a receptor like kinase called filagellin sensing 2 (FLS2) recognizes bacterial flagellin and leads to closure of stomata [28,30]. Similarly, the COR-deficient mutant of P. syringae pv. tomato (Pst) causes less disease when inoculated on the leaf surface than when inoculated directly into the apoplast of Arabidopsis or tomato [28].
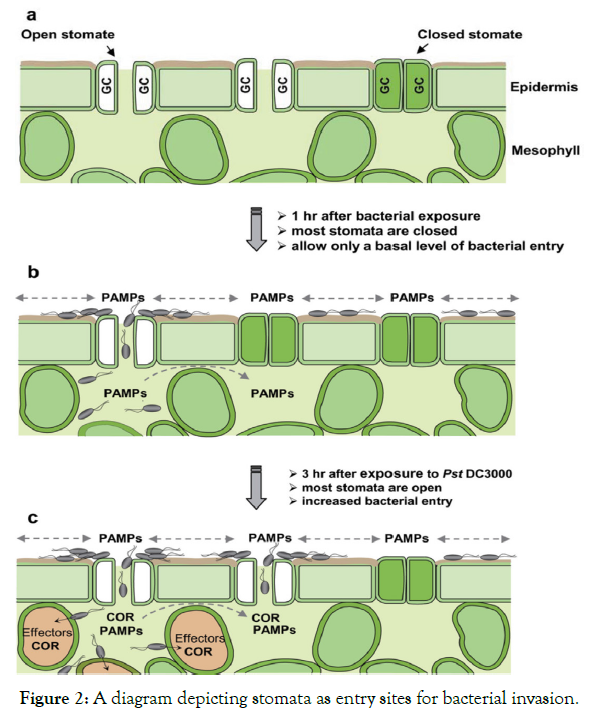
Figure 2: A diagram depicting stomata as entry sites for bacterial invasion.
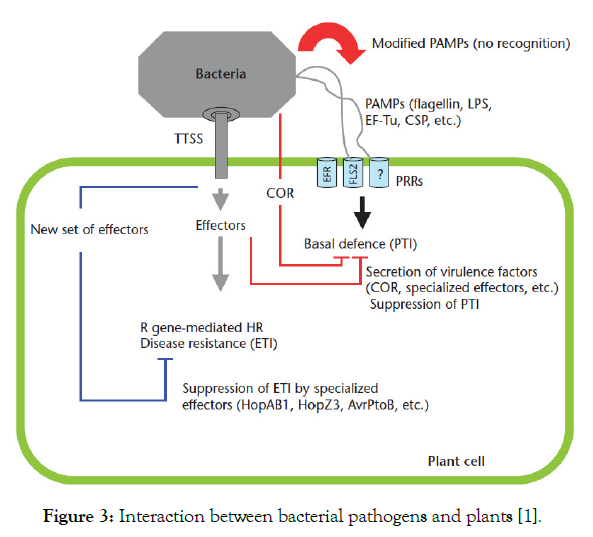
Figure 3: Interaction between bacterial pathogens and plants [1].
Generally, stomata open and close daily, reflecting the internal circadian rhythm of plants. However, bacteria can trigger stomatal closure under bright daylight [31], suggesting that stomatal guard cells can perceive bacteria and trigger a signaling cascade that overrides the natural circadian rhythm of stomatal movement. In other words, the foundation for a direct demonstration that guard cells surrounding the stomatal pore can sense microbes and close the pore, a process that is now known as stomatal defense or stomatal immunity [30]. Bacterium-triggered stomatal closure is a fast response (<1 hr.). Flagellin recognition has a prominent role in stomatal defense during the Arabidopsis-P. syringae pv. tomato DC3000 interaction [32] the existence of other PAMP-PRR pairs that function in stomatal defense is likely. In other words, the FLS2mutant of Arabidopsis, which lacks the receptor for bacterial flagellin, is more susceptible than the wild-type plant only when surface-inoculated with Pst DC3000 (Figure 2).
The wax layer and cuticle: Waxes are mixture of long chain aliphatic compounds which prevent the retention of water on plant surface essential for microbial growth. Because the epidermal cells of aerial plant parts are often covered in a waxy cuticle that is made up of fatty acids, leaf surfaces are always negatively charged. This charge often repels airborne inoculums of several microbes from settling on them. Thus in addition to prevention of water loss from the plant, the fatty acid layer also prevents microbial patho gens from coming into direct contact with epidermal cells thereby limiting infection [33]. The hydrophobic nature of the cuticle also prevents water from collecting on the leaf surface, an important defense against many pathogens that require standing water on the leaf surface for germination and multiplication [34].
Epidermal layer: Epidermis is the first layer of living host cells that comes in contact with attacking microbes. It is the first line of defense against invading pathogens and consists of both specialized and unspecialized cells. The toughness of epidermis is due to the polymers of cellulose, hemicelluloses, lignin mineral substances, polymerized organic compounds, suberin etc. Suberization of epidermis confers protection against plant X. axonopodis pv. Citri because of broad cuticular lips covering the stomata [4].
The plant epidermis is covered by a waxy cuticle and each cell is surrounded by a complex cell wall containing highly cross-linked polysaccharides, proteins and phenolic compounds that bacterial pathogens must overcome to access cell nutrients. To breach these barriers, many plant pathogenic bacteria produce a range of extracellular virulence factors such as cutin-degrading enzymes and cell wall-degrading enzymes such as cellulases, pectinases and endoglucanases. Such enzymes are particularly important in causing soft-rot diseases induced by bacteria in the Erwinia genus now a day pectobacteria [1].
Cell wall: If pathogens can pass the cuticle or enter through stomata they have to face another formidable obstacle, the cell wall. The cell wall is a major line of defense against bacterial pathogens providing an excellent structural barrier [35]. Structurally, it incorporates a wide variety of chemical defenses that can be rapidly activated when the cell detects the presence of potential pathogens. All plant cells have a primary and secondary cell wall, with the former giving structural support and is essential for turgor pressure, and the later developing inside of the primary cell wall after the cell’s active growth has stopped. Structurally, the primary cell wall is made of mainly cellulose, a complex polysaccharide consisting of thousands of glucose monomers linked together to form long polymer chains. These chains are bundled into fibers called micro fibrils, which give strength and flexibility to the wall [4].
The cell wall may also contain two groups of branched polysaccharides: cross-linking glycans and pectins. Cross-linking glycans include hemicellulose fibers that give the wall strength via cross-linkages with cellulose. Pectins form hydrated gels that help “cement” neighboring cells together and regulate the water content of the wall. Soft-rot pathogens often target pectins for digestion using specialized enzymes that cause cells to break apart causing brown and “mushy” appearance in fruits or vegetables [4]. Several species of bacteria colonize the spaces between cells (apoplast), and interaction with the cell wall is required to overcome defense responses and achieve full pathogenicity.
Pre-existing biochemical defense
Plants liberate different chemicals, which interfere with activities of the pathogen and pathogenesis, thereby preventing or reduce infection. These chemicals and the biochemical conditions that develop may act either directly through toxic or lytic effect on the invader or indirectly through stimulating antagonistic plant surface microflora. The compounds pre-existing in plants as constitutive antibiotics and those, which are formed in response to wounds as wounds antibiotics [4]. Chemical defenses include various antimicrobial peptides, proteins, and non-proteinaceous secondary metabolites present in plant cells that can prevent entrance of the invader [36]. Besides being directly harmful to the invader, they can operate by inactivating the extracellular enzymes secreted by the pathogen [37].
Anti-microbial compounds: Plants while growing and developing release gases as well as organic substances, from leaves and roots (leaf and root exudates), containing sugars, amino acid, organic acids, enzymes, glycoside etc. These materials have profound effect on the nature of surrounding environment, particularly the phyllosphere, rhizosphere microflora and fauna. Although these substances are ideal nutrients for microbes and help in germination and growth of several saprophytes and parasites, number of inhibitory substances is also present in these exudates. These inhibitory substances directly affect the microorganism or encourage certain groups to dominate the environment and function as antagonists of the pathogen [12].
Terpenoids such as of the monoterpenoids and sesquiterpenoids which are primary components of essential oils, are highly volatile compounds that contribute to the fragrance (essence) of plants that produce them. Essential oils often protect against bacterial attack. Mint plants (Mentha spp.) produce large quantities of the monoterpenoids menthol and menthone which are produced and stored in glandular trichomes on the epidermis. Examples of such terpenoids and their sources include peppermint and spearmint (Mentha spp.), basil (Ocimum spp.), oregano (Origanum spp.), rosemary (Rosmarinus spp.), sage (Salvia spp.), savory (Satureja spp.), thyme (Thymus spp.), black pepper (Piper spp), cinnamon (Cinnamomum spp), and bay leaf (Laurus spp) [4].
Diterpenoidsgossypol produced by cotton (Gossypium hirsutum) has strong antibacterial properties.Limonoidtriterpenoids responsible for the fresh scent of lemon and orangepeels, azadirachtin from the neem tree (Azadirachta indica) andcitronella from lemon grass (Cymbopogon citratus) have antibacterial activities even in low concentrations as few partsper million [38]. Saponins are glycosylated triterpenoids (triterpenoids with attached sugar groups) that are present in the cell membranes of many plant species. They have detergent (soap-like) properties and act by disrupting the cell membranes of invading bacterial pathogens [4].
Alkaloids are a large class of bitter-tasting nitrogenous compounds that are found in many vascular plants and include caffeine, cocaine, morphine, and nicotine. Caffeine and theobromine are alkaloids found in plants such as coffee (Coffea arabica), tea (Camellia sinensis), and cocoa (Theobroma cacao). They are toxic to microbes. In fact, high levels of caffeine produced by coffee seedlings can even inhibit the germination of other seeds in the vicinity of the growing plants, a phenomenon called allelopathy. Allelopathy allows one plant species to “defend” itself against other plants that may compete for growing space and nutrient resources. Members of the nightshade family (Solanaceae) produce many important alkaloid compounds. Nicotine is an alkaloid that is produced in the roots of tobacco plants (Nicotiana tabacum) and transported to leaves where it is stored in vacuoles [4,12].
Phytohormones: Production of the phytohormones auxins (e.g. indole-3-acetic acid-IAA) and cytokinins are important virulence factors for the gall-forming phytopathogenic bacteria, Pantoea agglomeranspv. gypsophilae, which causes crown and root gall disease of Gypsophila paniculata, and the pvs. savastanoi and nerii of Pseudomonas savastanoi, which causes olive and oleander knot disease [18,39].
Phytohormones also play a very important role systemic signaling that facilitates resistance against phytopathogens. For instance, a balance between salicylic acid, a local and systemic signal for resistance against many biotrophs, and the combination of jasmonic acid and ethylene accumulation sustain signals that promote defense against necrotrophs [18,40].
Induced defense mechanisms against bacterial pathogens
On the top of pre-existing defense mechanisms, plants develop an active/induced defense strategies when bacterial pathogens inter to the plant system like: biochemical defense, histological defense (lignification, suberization, abscission layers, tyloses, gum deposition, etc.) and induced cellular defense [1,4].
Inducible plant defenses are triggered by the perception of a pathogen or pathogen-derived molecules called elicitors. The elicitors can be either general, common to a group of microbes, or specific to certain pathogen strains. The induced biochemical changes in host plants are the last line of host defense. This may condition a plant or plant tissue from susceptible to resistant to immune status as per their genetic potential. The role of biochemical factors in host defense is based on four main attributes which include, a) association of the substance protection against disease at the site where protection occurs, b) isolation of the substance from the host showing protection against the disease, c) conferment of protection by the isolated substance when introduction into to the appropriate susceptible host and, d) resemblance of the nature of protection induced to the natural agents of a resistant plant [4,41].
During the initial contact between a plant and a bacterial pathogen, the plant can detect bacterial PAMPs such as flagellin and lipopolysaccharides (LPS). The perception of PAMPs activates signal- transduction cascades that turn on basal defenses. These basal defense responses include callose and silicone deposition to reinforce the cell wall, production of ROS and ethylene, transcriptional induction of a large suite of defense genes, including PR genes and post-transcriptional suppression of the auxin-signalling pathway. These responses are triggered by plant extracellular receptors specialized in the recognition of PAMPs termed PRRs (Figure 3). These basal defenses are generally sufficient to halt the growth of pathogenic and nonpathogenic microbes and prevent their establishment [1].
There are two major layers of biochemically induced plant immunity. These comprises; PTI and ETI. PTI is initiated by the perception of microbe conserved PAMPs, such as bacterial flgellin, LPS and elongation factor thermo unstable (EF-Tu), with specific plant cell surface pattern-recognition receptors (PRRs). PTI usually activates some early resistance responses, including stomatal closure, activation of mitogen-activated protein kinase (MAPK) cascades, transcription of resistance-related genes, ROS production and callose deposition. ETI on the other hand thought to a form of accelerated and amplified PTI response. It is activated by plant intracellular R genes (proteins) after specific perception of pathogenic type three secreted effectors (T3SEs), and is associated with programmed cell death, a response which is referred to as the HR [20,42].
Pathogen associated molecular patterns (PAMPS): Plant bacterial pathogens, and in general other pathogens, reveal themselves to the host immune system through molecules called PAMPs, such as flagellin or bacterial LPS and peptidoglycan. Since non-pathogenic bacteria also have these structures, PAMPs are also referred to as MAMPs [25,43]. PAMP-induced innate immunity takes place at the early stages of bacterial attack. PAMPs are usually recognized directly by receptors called plasma-membrane-localized PRRs which contain a leucine-rich repeat (LRR) domain outside of the plasma membrane and a cytoplasmic kinase domain [9] (Figure 3).
The best studied PAMP is flagellin. It is the principal component of bacterial flagellum, and is present in large amounts on nearly all flagellated bacteria. Flagellin, the major protein component of bacterial flagella as well as a well characterized PAMP, has been shown to be recognized by the Leu-rich repeat receptor (LRR) kinase FLS2 in Arabidopsis [44]. This recognition triggers a series of defense responses such as the MAPK cascades and the inhibition of multiplication of the pathogen. The response includes closure of stomata, which restricts bacterial penetration. FLS2, located in the plasma membrane, is believed to be involved in early bacterial-plant interaction by recognizing and binding flagellin. Flagellin and flg22 are detected in Arabidopsis by the PRR FLS2, which is a trans-membrane receptor kinase. Arabidopsis FLS2 mutants are more susceptible to infection by P. syringae pv. tomato DC3000 (Pst DC3000) when the bacteria are applied to the leaf surface, but not when the bacteria are infiltrated into the leaf intercellular space [45]. This observation indicates that FLS2 activates early defense responses that restrict penetration of bacterial pathogens into the plant tissue. Consistent with this conclusion, the FLS2 protein is expressed in epidermal cells and stomatal-guard cells, as well as mesophyll cells and cells in the stem, flower petals and roots [1].
LPS is another well-known PAMP. LPS, a major component of the outer membrane of Gram-negative bacteria, activates basal defense responses in plants. It was reported that purified LPS of X. campestris pv. campestris induces the oxidative burst in tobacco cells [46]. The purified lipo-oligosaccharide of X. campestris pv. campestris strain 8004 induces PR-1 and PR-2 gene expression in Arabidopsis [47]. Melotto et al. [30] were reported that purified LPS of P. syringae pv. tomato DC3000 induces the stomata closure which block the entrance of bacteria.
In addition to flagellin and LPS, the EF-Tu is another well-studied plant PAMP/PRR pair that activates defense responses similar to those triggered by recognition of flg22. It is the most abundant protein in a growing bacterial cell, acts as an inducer of basal defenses in plants. EF-Tu is a 43 kilo-daltons (kDa) protein. The N-terminal 18 amino acids of EF-Tu, elf18, can trigger basal defenses by itself. Recognition of elf18 in Arabidopsis occurs through a receptor-like kinase (RLK) named EF-Tu receptor (EFR). EFR has a similar structure to FLS2, including an extracellular LRRs domain and an intracellular kinase domain [48]. Transformation of Nicotiana benthamiana, which is unable to perceive EF-Tu, with EFR confers the ability to respond to elf18. Likewise, Arabidopsis plants containing mutations in EFR are unable to respond to elf18. Such EFR mutants display enhanced susceptibility to Agrobacterium tumefaciens transformation, suggesting that EF-Tu is required for triggering plant defenses induced by Agrobacterium.
Another bacterial molecule that acts as a PAMP is the CSP, which induces defense responses in solanaceous plants such as tobacco and tomato. The response to this PAMP has not yet been found outside of the Solanaceae family, indicating that some PRRs have evolved relatively late in angiosperm evolution. CSPs are small highly conserved proteins (~7.4 kDa) found in eubacteria. In contrast to flagellin and EF-Tu, the receptor for CSP has not yet been identified [1].
Gene-specific resistance and effector-triggered immunity (ETI): Plant disease resistance often depends on specific recognition of the pathogen and a rapid induction of general defense responses. Usually such a recognition event is a gene-for gene interaction, depending on a R gene in the plant and its corresponding Avr gene in the pathogen. More than 40 plant disease Rgenes have been cloned in different plant species. These R genes confer resistance to diverse pathogens and can be classified based on their protein structures into several groups [49].
The recognition of an Avr protein by an R protein triggers a series of defense responses that enable the plant to protect itself and defeat the pathogen in order to survive. This detection system employs intracellular receptors encoded by ‘disease-resistance’ (R) genes. The majority of plant R proteins contain a nucleotide-binding site and LRR (NBS–LRR). NBS–LRR proteins mediate resistance against a large range of plant pathogens. Activation of an R protein by a pathogen effector protein typically leads to activation of programmed cell death of plant cells surrounding the pathogen. This localized cell death is referred to as the hypersensitive response (HR) and such R protein mediated resistance is referred to as effector-triggered immunity (ETI) [1].
At the molecular level, recognition of pathogen effectors by plant R proteins can either be direct or indirect (Figure 4). The direct recognition model, also termed the ligand receptor model, proposes that disease-resistant proteins function as receptors to bind pathogen-encoded effector proteins directly. Direct recognition has been observed for a bacterial pathogen, R. solanacearum, in which the R protein RRS1 binds directly to the R. solanacearum-effector PopP2 [50]. The indirect mode of recognition, also termed the ‘Guard model’, involves the detection of modifications made to host proteins by effectors. This mode of recognition seems to be common in bacterial pathogens and has been observed for the P. syringae effectors AvrPphB, AvrRpm1/AvrB, AvrRpt2 and AvrPtoB [51] (Figure 4).
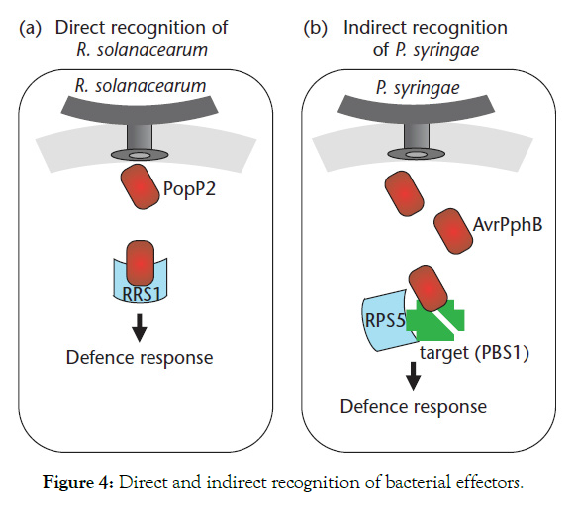
Figure 4: Direct and indirect recognition of bacterial effectors.
Reactive oxygen species (ROS): ROS are chemically reactive chemical species containing oxygen include hydrogen peroxide (H2O2), superoxide (O2-), hydroxyl radical (.OH-) and singlet oxygen (1O2) normally occurs in the metabolism of plant cells [52]. In a biological context, ROS are formed as a natural byproduct of the normal metabolism of oxygen and have important roles in cell signaling. They are usually generated by the electron transport activities of chloroplasts and mitochondria, and by enzymes in other cell compartments (sections) and the apoplast, involved in reduction-oxidation processes of the plant cell (Figure 5).
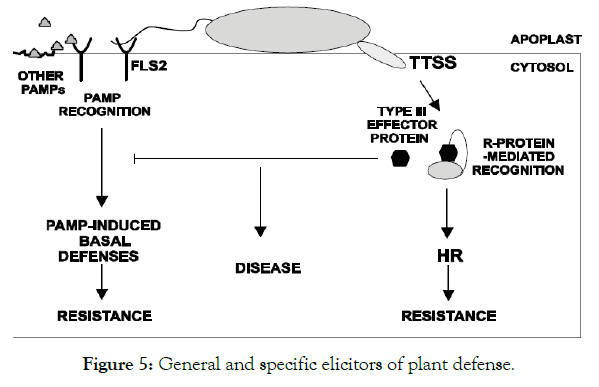
Figure 5: General and specific elicitors of plant defense.
When a plant recognizes an attacking bacterial pathogen, one of the first induced reactions is to rapidly produce O2- or H2O2 to strengthen the cell wall. This prevents the spread of the pathogen to other parts of the plant, essentially forming a net around the pathogen to restrict movement and reproduction. For example, Thiamine (vitamin B1) induces resistance against P. syringae pv.tomato DC3000 (PstDC3000), which is associated with H2O2- dependent priming of defense genes and callose deposition. Vitamin B2 (riboflavin) induces a phenotypically similar resistance response that is associated with priming of ROS production, callose deposition and SA-inducible genes [53]. The plant secondary metabolite quercetin has also been demonstrated to induce SA and non-expresser PR genes 1 (NPR1) dependent resistance against PstDC3000, which is associated with augmented deposition of ROS, callose, PR1and phenylalanine ammonia-lyase (PAL)gene transcripts [54]. A study by Mukherjee et al. [55] provided a plausible mechanism for ROS dependent regulation of priming. The authors performed a phenotypic analysis of different alleles of the ascorbic acid deficient mutant vtc1 and demonstrated that the enhanced disease resistance of this mutant is based on priming of pathogen induced accumulation of ROS, SA and NPR1gene transcripts [55]. The authors suggested that the reduced ROS scavenging capacity of vtc1causes constitutive priming of pathogen induced H2O2, thereby causing augmented SA accumulation and enhanced defense induction.
In early studies, Adam et al. [56] reported, tobacco plants infected with P. syringae pv. syringae exhibited an HR that was accompanied by increased O2- generation and lipid peroxidation. In tobacco cell suspensions treated with P. syringae, all bacterial treatments with resulted in an initial, rapid oxidative burst (0-1 hr.) as detected by the chemiluminescence assay [57]. In another study, a protein termed harpin has been identified as an elicitor of the HR in tobacco caused by E. amylovora. Harpin-producing E. amylovora induced active oxygen species (AOS) production in tobacco cell suspensions after a lag of 2 hr., but AOS were not induced by E. amylovora transposon mutants that do not produce harpin [58].
Systemic Acquired Resistance (SAR): Besides blocking pathogen replication and the spread of infection, ETI can also trigger a general defense response referred to as SAR [59]. This phenomenon is conserved among diverse plants and confers long-lasting resistance. During SAR systemic tissues are primed for defense, normally from weeks to months, thus protecting the whole plant from secondary infection [60] Also PTI induces a form of systemic resistance very similar to SAR. Thus, treatment with PTI-inducing P. syringae and ETI-inducing AvrRpm1 elicit a highly similar systemic response in Arabidopsis [61,62]. SAR generally leads to accumulation of metabolites like the small phenolic hormone SA, and enhanced systemic expression of a variety of classical defense and SAR marker genes such as pathogenesis-related genes 1 (PR1), 2 (PR2), and 5 (PR5) [61,62]. SA induces SAR related gene expression via the downstream regulator non-expresser of PR genes 1 (NPR1), a transcriptional co-activator and SA receptor [60]. The resulting signal thought not only to include codes for priming of defenses, but also encode detailed information about the primary pathogen infection. Intriguingly, studies on different plant species suggests that the composition of the mobile immune signal in SAR differs depending on the plant species and the type of pathogen conducting the primary infection [63] (Figure 6).
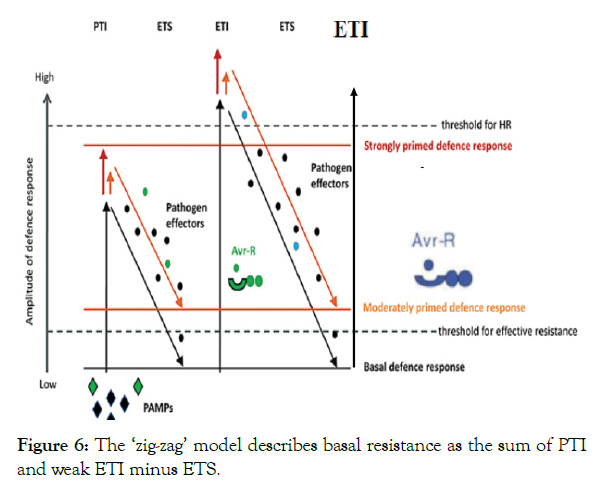
Figure 6: The ‘zig-zag’ model describes basal resistance as the sum of PTI and weak ETI minus ETS.
Cycles of PTI, ETS, and nod-like receptor (NLR)-mediated ETI are major forces shaping plant host-pathogen coevolution and have led to the so called zigzag model (Figure 6) [64]. In this model, pathogen effectors are like double-edged swords, triggering ETI in plants with the corresponding NLRs on the one hand, but exhibiting virulence activities in plants in the absence of the corresponding NLRs on the other hand. This co-evolution of plants and pathogens over millions of years has culminated in large arsenals of immune receptors present in plant genomes [65].
In general, the zigzag model (Figure 6) can be summarized in four main stages. 1: plants detect MAMPs via PRRs to trigger PAMP-triggered immunity (PTI). 2: successful pathogens deliver effectors that interfere with PTI, resulting in effector triggered susceptibility (ETS). 3: an effector can be recognized by an NB-LRR protein, activating ETI, which after surpassing a defined threshold induces hypersensitive cell death (HR). 4: pathogen strains that have lost certain effector are selected. They might have also gained a new set of effectors to respond to the plant defense.
Phytoalexins: Phytoalexins are low molecular weight antimicrobial compounds that are produced by plants as a response to biotic and abiotic stresses [66]. They are synthesized by either the cells adjacent to the infection site, the infected host cells or by the invading pathogen. It is thought that such infected cells produce some sort of signals which induces the adjacent cells to produce the phytoalexins, which are packaged in lipid vesicles and exported to the infected cell/site [4]. Consequently, the infected cell becomes a toxic micro-environment for the invading pathogen. The rapidity of phytoalexin accumulation is associated with resistance in plants to diseases caused by bacteria, although the genetic information for phytoalexin synthesis is found in susceptible and resistant plants [66]. Phytoalexin accumulation is often associated with hypersensitive cell death. Examples include medicarpin (alfalfa, Medicago sativa), rishitin (tomatoes and potatoes (the Solanaceae family), and camalexin, (Arabidopsis thaliana) [18]. Phytoalexins are only one component of the complex mechanisms for disease resistance in plants.
Phytohormones as a defense signaling molecules: Phytohormones are signal molecules produced within the plant, and occur in extremely low concentrations. They are not only instrumental in regulating developmental processes in plants but also play important roles for the plant’s responses to biotic and abiotic stresses. It includes ethylene (ET), jasmonic acid (JA), SA, auxin and ABA. The molecular mechanisms that govern these hormonal networks are largely unknown. Moreover, hormone signaling pathways are targeted by pathogens to disturb and evade plant defense responses [67,68].
To develop hormone-based breeding strategies aiming to improve crop resistance to pathogens, we need to understand the intricate regulation of hormone homeostasis during plant-pathogen interactions, and how pathogens interfere with this hormone regulation. Indeed, manipulation of a plant hormone pathway can result in enhanced resistance to a particular pathogen, but it could also have a strong negative effect on plant growth and resistance to a distinct type of pathogen with a different life style [69].
Auxins are a group of molecules including IAA that regulate many aspects of plant development, such as apical dominance, root gravitropism, root hair, lateral root, leaf, and flower formation, and plant vasculature development [70]. Both direct and indirect effects of auxins on the regulation of pathogen resistance responses in plants have been described. Indirect effects may be caused by auxins regulation of development-associated processes, such as cell wall architecture, root morphology, and stomata pattern. For example, treatment of rice with IAA impaired the resistance to X. oryzae pv. oryzae probably as a consequence of the activation of the biosynthesis of cell wall-associated expansins that lead to cell wall loosening, which facilitates pathogen growth [71].
Auxins can negatively impact plant defense by interfering with other hormone signaling pathways or with PTI [72]. The bacterial PAMP flg22, a peptide from flagellin protein [73], induces an Arabidopsis microRNA (miR393), which negatively regulates the mRNA levels of auxins receptors TIR1 (transport inhibitor response 1), AFB2 (auxin signaling F-box 2), and AFB3. Thus, the flg22-triggered suppression of auxin signaling leads to increased resistance to the bacterium P. syringae pv. tomato DC3000 (PstDC3000). The flg22-induced resistance to these biotrophic pathogen was explained by the observed induction of the SA signaling pathway. Supporting this hypothesis, it was found independently that treatment of Arabidopsis leaves with flg22 induces SA accumulation [74].
Interaction between SA and auxins was further clarified by the characterization of the regulatory pattern of GH3.5 gene, which is involved in auxin homeostasis in Arabidopsis plants. Lines overexpressing GH3.5 have lower levels of Aux/IAA proteins, overexpression of SA signaling pathway and enhanced resistance to P. syringae [75]. The conjugated auxin aspartic acid (IAA–Asp) has been reported to play a key role in regulating resistance to PstDC3000. In Arabidopsis, tomato, and Nicotiana benthamiana infected with these pathogens there is an enhanced expression of GH3.2 and GH3.4 genes, which encode two enzymes required for conjugation of auxins with Asp. Thus, upon pathogen infection, accumulation of IAA–Asp takes place, promoting the development of disease symptoms in infected plants [76].
One of the biosynthetic pathways of auxins is partially shared with those required for the biosynthesis of tryptophan-derived antimicrobials, such as indole glucosinolates and camalexin. This might lead to competition for the biosynthetic precursor of auxin and antimicrobials [77]. The recently characterized Arabidopsis wat1 (walls are thin1) mutant exhibits specific enhanced resistance to vascular pathogens such as R. solanacearum. This response was associated to a miss-regulation of tryptophan derivatives (i.e., lower levels of auxin and indole glucosinolates) specifically in roots, resulting in enhanced levels of SA which is, like tryptophan, a chorismate-derivative [78]. Collectively, these data demonstrate that auxins play a central role in balancing plant resistance responses.
The function of SA in activating resistance against pathogens has been thoroughly described. In Arabidopsis, SA is synthesized from chorismate (a precursor of tryptophan and, consequently, of auxins) via two pathways, either through phenylalanine or through isochorismate [79]. This second pathway, in which SID2/ICS1 (salicylic acid induction deficient 2/isochorismate synthase 1) is involved, is activated upon pathogen infection, such as P. syringae, and after plant recognition of pathogen effectors or PAMPs [79]. Deficiency of SA biosynthesis in sid2-1 mutant leads to reduced resistance response in Arabidopsis plants. SA is a regulator of plant resistance to biotrophic and hemibiotrophic pathogens, such as P. syringae, and it also regulates SAR, a well-studied type of induced resistance [80].
NPR1 a well-known central player in SA signaling, and NPR3 and NPR4 proteins have been described as SA receptors [81,82]. NPR1 localizes at the cytosol as an oligomer, and in the presence of SA, redox changes occurs in NPR1 that lead to the dissociation of NPR1 complex and to the translocation of the corresponding monomers to the nucleus. There, NPR1 protein activates the transcription of defensive genes, such as PR protein, by interacting with TGA (TGACG sequence-specific binding protein) transcription factors [72]. Transcriptional regulation of SA-defensive genes is also mediated by HDA19 (histone deacetylase19) that repressed SA-mediated basal defense to PstDC3000 [83]. Up-regulation of SA marker genes (PR1, PR2, ICS1, EDS1) and over-accumulation of SA take place in HDA19 mutant, which correlates with its enhanced resistance phenotype to PstDC3000 pathogenic bacteria. Indeed, HDA19 targets PR1 and PR2 promoters to regulate gene expression. The mutation HDA19 causes hyper-acetylation of histones in the promoters of PR genes and priming of SA-associated plant defense [83].
ABA is an isoprenoid compound that regulates developmental processes, such as seed development, desiccation, and dormancy [84]. It can function as a positive or a negative regulator of plant defense depending on the plant–pathogen interaction analyzed [85]. ABA-impaired (biosynthesis or signaling) mutants in tomato and Arabidopsis (abi1-1, abi2-1, aba1-6, aba2- 12, aao3-2, and pyr1pyl1pyl2pyl4) were shown to overexpress defensive-signaling pathways, leading to enhanced resistance to different pathogens such as P. syringae [86]. Negative interactions of ABA with the major hormones involved in plant defense response (SA, JA, and ET) have been described by means of exogenous hormone treatments [86]. At the pre-invasion stage, ABA promotes resistance to bacterial infection as it favors stomatal defense [87,88]. For instance, purified MAMP and live Pst DC3000 do not induce stomatal closure in the ABA-deficient aba3-1 mutant [30]. Similarly, the notabilis mutant of tomato, which lacks a functional ABA biosynthesis enzyme, 9-cis-epoxycarotenoid dioxygenase, is also compromised in Pst DC3000-induced stomatal closure [89]. Furthermore, the core signaling components of the ABA pathway that lead to stomatal closure in Arabidopsis are involved in Pst-triggered stomatal closure. In particular, these components include:- pyrabactin resistance1/pyrabactin resistance1-like/regulatory components of ABA receptors; protein phosphatase 2CA; OPEN STOMATA1 (OST1); the ABA signaling-related secondary messengers ROS, NO, Ca2+, and G-protein α-subunit; and the membrane channels SLOW ANION CHANNEL-ASSOCIATED1 (SLAC1) and K+ channels [28,30,90]. Thus, current experimental evidence suggests a prominent role of ABA signaling in stomatal defense [87,88].
ABA and SA have been shown to function antagonistically in the control of the resistance to some pathogens, they trigger stomata closure to avoid penetration of the bacteria P. syringae in Arabidopsis [30] Plant treatment with flg22 is known to interfere with ABA signaling to induce stomata closure. The ABA- or flg22-induced stomata closure are impaired in lines overexpressing HSC70-1 (heat shock cognate70-1) and mutants in HSP90 (heat shock protein90) [91], resulting in an increased susceptibility to both virulent and avirulent strains of P. syringae [92]. ABA is also a key hormone in Arabidopsis response to R. solanacearum infection, as 40% of the genes up-regulated during the development of wilting symptoms were related to ABA, including those encoding proteins for ABA biosynthesis [i.e., 9-cis-epoxycarotenoid dioxygenase3 (NCED3)] or signaling [i.e., ABA-insensitive1 (ABI1) and ABI5] [93]. More recently, it has been shown that pre-inoculation of Arabidopsis with an avirulent strain of R. solanacearum activates plant resistance to virulent isolates of this bacterium, and this resistance was correlated with the enhanced expression of ABA-related genes that resulted in a hostile environment for the infection development. These results also suggest that ABA may be used in biological control of bacterial wilt caused by R. solanacearum [94].
Several derivatives of jasmonates (JAs) are naturally present in plants, some of which are biologically active for regulating JA-associated biological responses. It is also an important signaling molecule for the activation of defense in response to pathogen attack. Indications for a role of JA for pathogen defense in potato arose from reports that exogenous application of JA leads to local and systemic protection against subsequent pathogen attack. Moreover, in response to infection with the non-host bacteria P. syringae pv. maculicola or in response to treatment with the oligopeptide elicitor Pep-13, infiltrated potato leaves accumulate not only salicylic acid, but also JA [67].
The role of enzymes and pathogenesis related proteins as defense mechanisms: As it was discussed above, in general, plant pathogens reveal themselves to a host’s immune system through molecules called PAMP or MAMP, such as flagellins or bacterial lipopolysaccharides. Abundant proteins and enzymes are found in plants mainly in plant cell walls that actively work to reshape the wall during cell growth and also thicken and strengthen the wall during induced defense. In addition, their physiological function is strong antimicrobial activity, but they also display lysozymal activity and thus may be involved in conferring resistance to plants against bacterial pathogens Post-inflectional changes in host cells involve production and modification of large number of proteins (structural and enzymatic), which have important role in defense mechanism. The enzymes are required for various synthetic pathways (normal or modified) for production of resistance related substances. Some plants and seeds also contain proteins and enzymes that specifically inhibit pathogens [4]. However, unlike phytochemicals such as terpenoids, phenolics, and alkaloids, proteins require a great deal of plant resources and energy to produce; consequently, many defensive proteins are only made in significant quantities after a pathogen has attacked the plant. Once activated, however, defensive proteins and enzymes effectively inhibit bacteria. Some of the proteins and enzymes include, defensins, amylase inhibitors, lectins, ricin, protease inhibitors, hydrolytic enzymes, chitinases, and lucanases, peroxidases, and polyphenol oxidases, lysozymes and hydroxyproline-rich glycoproteins [41,95] (Figure 7 and Table 3).
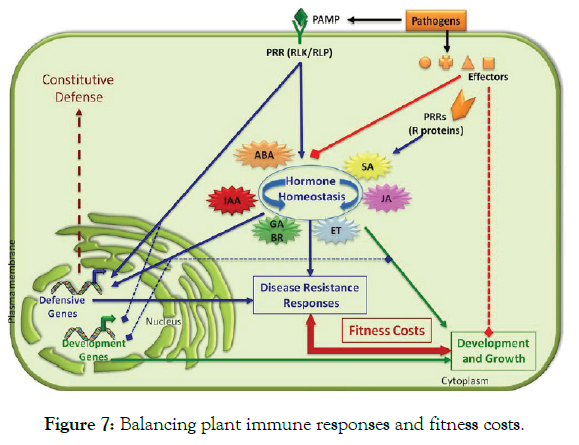
Figure 7: Balancing plant immune responses and fitness costs.
| Families | Type member | Properties |
|---|---|---|
| PR-1 | Tobacco PR-1a | Antifungal |
| PR-2 | Tobacco PR-2 | β-1,3-glucanase |
| PR-3 | Tobacco P, Q | Chitinase type I, II, IV, V, VI, VII |
| PR-4 | Tobacco ‘R’ | Chitinase type I,II |
| PR-5 | Tobacco S | Thaumatin- like |
| PR-6 | Tomato Inhibitor I | Proteinase- inhibitor |
| PR-7 | Tomato P69 | Endoproteinase |
| PR-8 | Cucumber chitinase | Chitinase type III |
| PR-9 | Tobacco ‘lignin forming peroxidase’ | Peroxidase |
| PR-10 | Parsley ‘PR1’ | Ribonuclease like |
| PR-11 | Tobacco ‘class V’ chitinase | Chitinase, type I |
| PR-12 | Radish Rs- AFP3 | Defensin |
| PR-13 | Arabidopsis THI2.1 | Thionin |
| PR-14 | Barley LTP4 | Lipid- transfer protein |
| PR-15 | Barley OxOa (germin) | Oxalate oxidase |
| PR-16 | Barley OxOLP | Oxalate oxidase-like |
| PR-17 | Tobacco PRp27 | Unknown |
Table 3: Classification of pathogenesis related proteins.
Proteins encoded by the host plant induced under pathological or related conditions are termed pathogenesis-related (PR) proteins. PR protein was first discovered and reported in tobacco plants infected by tobacco mosaic virus [96]. Later, these proteins were found in many plants. Most PR proteins in the plant species are acid-soluble, low molecular weight, and protease-resistant proteins virus [96]. PR proteins depending on their isoelectric points may be acidic or basic proteins but they have similar functions. Most acidic PR proteins are located in the intercellular spaces, whereas, basic PR proteins are predominantly located in the vacuole [97]. PR-proteins were categorized into 17 families according to their properties and functions (Table 3). Among these PR proteins chitinases and β-1, 3-glucanases are two important hydrolytic enzymes that are abundant in many plant species after infection by different type of pathogens. This co-induction of the two hydrolytic enzymes has been described in many plant species, including pea, bean, tomato, tobacco, maize, soybean, potato, and wheat.
Conclusion and Future Perspectives
Perception of bacteria by plant cells and activation of the basal defense response do not depend on a single bacterial factor but can be triggered by several different factors. Plants rely on the perception of multiple PAMPs for efficient recognition of bacterial pathogens. Since the activation of basal defense responses depends on the perception of PAMPs, it is also called PTI. To better understand the complex interactions between plants and bacterial pathogens, the field must continue to unravel the relative contributions of PRR-mediated and R-protein-mediated resistance to promoting plant immunity, and the role of PAMP variation and effector virulence activity in avoiding or suppressing plant defenses. Eventually, it might be possible to predict the outcome of a given plant–bacterium interaction by simply knowing the complete complement of PAMPS, effectors, PRRs and R proteins that are in the system. It is possible that the observed differences between PRR-mediated and R-protein-mediated resistance might be due to the strength or timing of defense response elicitation or the relative recalcitrance to suppression by type III effectors. In practical terms, it might be useful to redefine plant defenses as early, extracellular defenses (PRRs) and later, intracellular defenses (R proteins). As the distinctions between PRR-mediated and R-protein-mediated defenses blur, it will be interesting to test to what degree the responses are shared between these plant defenses.
Identifying effector targets will help us to understand both bacterial pathogenesis and host resistance. How R proteins are activated by effectors to induce defense responses, including the HR, is also needs further investigation. Finally, the biggest challenge for the future is applying our increasing knowledge of the plant immune system to development of more bacterial-resistant crops, and in particular, development of crops that display durable resistance across time and space. Identification of key effector targets, and the R proteins that guard them, may facilitate development of such crops.
REFERENCES
- Ade J, Innes R. Resistance to bacterial pathogens in plants. Encyclopedia of Life Sciences, John Wiley & Sons, Ltd. 2007.
- Agrios GN. Plant Pathology (5th edn). Academic Press, San Diego, CA, USA. 2005.
- Starr MP. Bacteria as plant pathogens. Ann Rev Microbiol. 1959;13(1):211-38.
- Doughari JH. An Overview of Plant Immunity. J Plant Pathol Microbiol. 2015;6:322.
- Velasquez AC, Oney M, Huot B, Xu S, He SY. Diverse mechanisms of resistance to Pseudomonas syringae in a thousand natural accessions of Arabidopsis thaliana. New Phytologist. 2017;214:1673–1687.
- Musidlak O, Buchwald W, Nawrot R. Plant defense responses against viral and bacterial pathogen infections. Focus on RNA-binding proteins (RBPs). Kerla Korlonica. 2014;60:1-4
- Kannan VR, Bastas KK. Sustainable approaches to controlling plant pathogenic bacteria. CRC; 2015;8.
- Teper D, Sunitha S, Martin GB, Sessa G. Five Xanthomonas type III effectors suppress cell death induced by components of immunity-associated MAP kinase cascades. Plant Signal Behavior. 2015;10:1064573.
- Abramovitch RB, Anderson JC, Martin GB. Bacterial elicitation and evasion of plant innate immunity. Nature Reviews Mol Cell Biol. 2006;7(8):601-611.
- Buttner D, He SY. Type III Protein secretion in plant pathogenic bacteria. Plant Physiol. 2009;150:1656–1664.
- Schechter LM, Roberts KA, Jamir Y, Alfano JR, Collmer A. Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J Bacteriol. 2004;186:543–555.
- Buonaurio R. Infection and plant defense responses during plant-bacterial interaction. In: Barka, EA, Clément C (eds). Plant-Microbe Interactions. 2008;169-197.
- Lodha TD, Hembram P, Tep N, Basak J. Proteomics: A successful approach to understand the molecular mechanism of plant-pathogen interaction. Am J Plant Sci 2013,4:1212-1226.
- Mehta A, Rosato YB. Differentially expressed proteins in the interaction of Xanthomonas axonopodis pv. citri with leaf extract of the host plant. Proteomics. 2001;1:1111–1118.
- Tahara ST, Mehta A, Rosato YB. Proteins induced by Xanthomonas axonopodis pv. passiflorae with leaf extract of the host plant (Passiflorae edulis). Proteomics. 2003;3:95–102.
- Cao X, Fan G, Dong Y, Zhao Z, Deng M, Wang Z, et al. Proteome profiling of Paulownia seedlings infected with phytoplasma. Front Plant Sci. 2017;8:342.
- Jones AME, Thomas V, Truman B, Lilley K. Mansfield J, Grant M. Specific changes in the Arabidopsis proteome in response to bacterial challenge: differentiating basal and R-gene mediated resistance. Phytochem 2004;65:1805–1816.
- Jones AME, Thomas V, Bennett MH, Mansfield J, Grant M. Modifications to the Arabidopsis defense proteome occur prior to significant transcriptional change in response to inoculation with Pseudomonas syringae. Plant Physiol. 2006;142:1603–1620.
- Suárez-Moreno ZR, Caballero-Mellado J, Coutinho BG, Mendonça-Previato L, James EK, Venturi V. Common features of environmental and potentially beneficial plant-associated Burkholderia. Microbial Ecol. 2012;63:249-266.
- Mahmood T, Jan A, Kakishima M, Komatsu S. Proteomic analysis of bacterial-blight defence responsive proteins in rice leaf blades. Proteomics. 2006;6:6053–6065.
- Chen F, Yuan Y, Li Q, He Z. Proteomic analysis of rice plasma membrane reveals proteins involved in early defense response to bacterial blight. Proteomics. 2007;7:1529–1539.
- Mathesius U, Mulders S, Gao M, Teplitski M, Caetano- Anolles G, Rolfe, BG, et al. Extensive and specific responses of a eukaryote to bacterial quorum-sensing signals. Proceedings of the National Academy of Sciences. 2003;100:1444–1449.
- Jibril SM, Jakada BH, Kutama AS, Umar HY. Plant and Pathogens: Pathogen Recognision, Invasion and Plant Defense Mechanism. International Journal of Current Microbiology and Applied Sciences. 2016;5(6):247-257.
- Mackey D, McFall AJ. MAMPs and MIMPs: proposed classifications for inducers of innate immunity. Molecular Microbiol. 2006;61:1365-71.
- High Impact. Plant Response to Bacterial Pathogens. Overlap between Innate and Gene-for Gene Defense Response. Plant Physiol. 2006;142:809–811.
- Gudesblat GE, Torres PS, Vojnov AA. Stomata and Pathogens: Warfare at the Gates. Plant Signaling & Behavior. 2009;4(12):1114–1116.
- Underwood W, Melotto M, He SY. Role of plant stomata in bacterial invasion. Cellular Microbiol. 2007;9(7):1621–1629.
- Melotto M, Zhang L, Oblessuc PR, He SY. Stomatal defense a decade later. Plant Physiol 2017;174:561–571.
- Zipfel C. Plant pattern-recognition receptors. Trends Immunol. 2014;35:345–351.
- Melotto M, Underwood W, Koczan J, Nomura K, He SY. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126:969-980.
- Roy D, Panchal S, Rosa BA, Melotto M. Escherichia coli O157:H7 induces stronger plant immunity than Salmonella enterica Typhimurium SL1344. Phytopathol. 2013;103:326–332.
- Zeng W, He SY. A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiol. 2010;153:1188–1198
- Serrano M, Coluccia F, Torres M, L'Haridon F, Métraux JP. The cuticle and plant defense to pathogens. Front Plant Sci. 2014;5:274.
- Belete T, Boyraz N. Critical Review on Apple Scab (Venturia inaequalis) Biology, Epidemiology, Economic Importance, Management and Defense Mechanisms to the Causal Agent. J Plant Physiol Pathol. 2017;5:1-2.
- Great O, Nnenna G, Emma U. Defense Mechanisms in Plants against Invading Plant Pathogenic Microbes in Nigeria. J Agri Sustain. 2017;10:80-96.
- Nürnberger T, Brunner F, Kemmerling B, Piater L. Innate immunity in plants and animals: striking similarities and obvious differences. Immunol Rev. 2004;198:249–266.
- Zhao J, Davis LC, Verpoorte R. Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol Advances. 2005;23:283-333.
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323-329.
- Anderson JP, Gleason CA, Foley RC, Thrall PH, Burdon JB, Singh KB. Plants versus pathogens: an evolutionary arms race. Funct Plant Biol. 2010;37:499-512.
- Cai K, Gao D, Chen J, Luo S. Probing the mechanisms of silicon mediated pathogen resistance. Plant signal behaviour. 2009;4:1-3.
- Dou D, Zhou JM. Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Micro. 2012;12:484-495.
- Hou S, Yang S, Wu, D, Zhang C. Plant immunity: evolutionary insights from PBS1, Pto, and RIN4. Plant Signal Behavior. 2011;6:794-799.
- Ranf S, Gisch N, Schäffer M, Illig T, Westphal L, Knirel YA, et al. A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nature Immunol. 2015;16(4):426-433.
- Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell. 2006;18:465-476.
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JDG, Felix G, Boller T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767.
- Braun SG, Meyer A, Holst O, Puhler A, Niehaus K. Characterization of the Xanthomonas campestris pv. campestris lipopolysaccharide substructures essential for elicitation of an oxidative burst in tobacco cells. Mol Plant Microbe Interact. 2005;18:674-81.
- Silipo A, Molinaro A, Sturiale L, Dow JM, Erbs G, Lanzetta R, et al. The elicitation of plant innate immunity by lipooligosaccharide of Xanthomonas campestris. J Biol Chem. 2005;280:360-368.
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium- mediated transformation. Cell. 2006;125:749–760.
- Martin GB, Bogdanove AJ, Sessa G. Understanding the functions of plant disease resistance proteins. Ann Rev Plant Biol. 2003;54:23-61.
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, et al. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proceedings of the National Academy of Sciences. 2003;100:8024–8029.
- Ade J, DeYoung BJ, Golstein C, Innes RW. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proceedings of the National Academy of Sciences. 2007;104:2531–2536.
- Hayyan M, Hashim M, Al-Nashef IM. Superoxide Ion: Generation and Chemical Implications. Chem Rev. 2016;116(5):3029–3085.
- Zhang S, Yang X, Sun M, Sun F, Deng S, Dong H . Riboflavin-induced Priming for Pathogen Defense in Arabidopsis thaliana. J Int Plant Biol. 2009;51:167-174.
- Jia Z, Zou B, Wang X, Qiu J, Ma H, Gou Z, et al. Quercetin-induced H2O2 mediates the pathogen resistance against Pseudomonas syringae pv. Tomato DC3000 in Arabidopsis thaliana. Biochem Biophy Res Com. 2010;396:522-527.
- Mukherjee M, Larrimore KE, Ahmed NJ, Bedick TS, Barghouthi NT, Traw MB, et al. Ascorbic Acid Deficiency in Arabidopsis Induces Constitutive Priming That is Dependent on Hydrogen Peroxide, Salicylic Acid, and the NPR1 Gene. Mol Plant Microb Interact 2010;23:340- 351.
- Adam A, Farkas T, Somlyai G, Hevesi M, Kiraly Z. Consequence of O2- generation during a bacterially induced hypersensitive reaction in tobacco: deterioration of membrane lipids. Physiol Mol Plant Pathol. 1989;34:13-26.
- Mehdy MC. Active Oxygen Species in Plant Defense against Pathogens. Plant Physiol. 105:467-472.
- Baker CJ, Orlandi EW, Mock NM. Harpin, an elicitor of the hypersensitive response in tobacco caused by Erwinia amylovora, elicits active oxygen production is suspension cells. Plant Physiol. 1993;102:1341-1344.
- Mishina TE, Zeier J. Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J Cell Mole Biol. 2007;50:500-513.
- Fu ZQ, Dong X. Systemic acquired resistance: turning local infection into global defense. Ann Rev Plant Biol. 2013;64,839-863.
- Mishina TE, Zeier J. Pathogen‐associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. The Plant J. 2007;50(3):500-513.
- Navarova H, Bernsdorff F, Doring AC, Zeier J. Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. The Plant cell. 2012;24:5123-5141.
- Spoel SH, Dong X. How do plants achieve immunity? Defense without specialized immune cells. Nat Rev Immunol. 2012;12:89-100.
- Maekawa T, Kufer TA, Schulze-Lefert P. NLR functions in plant and animal immune systems: so far and yet so close. Nature Immunol. 2011;12:817-826.
- Toruno TY, Stergiopoulos I, Coaker G. Plant-Pathogen Effectors: Cellular Probes Interfering with Plant Defenses in Spatial and Temporal Manners. Ann Rev Phytopathol. 2016;54:419-441.
- Singh R, Chandrawat KS. Role of Phytoalexins in Plant Disease Resistance. Int J Cur Microbiol Appl Sci. 2017;6(1):125-129.
- Halim VA, Vess A, Scheel D, Rosahl S. The Role of Salicylic Acid and Jasmonic Acid in Pathogen Defense. Plant Biol. 2006;8:307–313.
- Denancé N, Sánchez-Vallet A, Goffner D, Molina A . Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front Plant Sci. 2013;4:1-12.
- Holeski LM, Jander GD, Agrawal AA. Trans-generational defense induction and epigenetic inheritance in plants. Trends Ecol Evol. 2012;27:618–626.
- Swarup R, Péret B. AUX/LAX family of auxin influx carriers – an overview. Front Plant Sci. 2012;3:221-225.
- Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, et al. Activation of the indole-3- acetic acid amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate independent basal immunity in rice. Plant Cell. 2008;20:228–240.
- Robert-Seilaniantz A, Grant M, Jones JD. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Ann Rev Phytopathol. 2011;49:317–343.
- Pel MJC, Pieterse CMJ. Microbial recognition and evasion of host immunity. J Exp Bot. 2012;64:1237–1248.
- Tsuda K, Sato M, Glazebrook J, Cohen JD, Katagiri F. Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 2008;53:763–775.
- Park JE, Park JY, Kim YS. Staswick PE, Jeon J, Yun J, et al. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J Biol Chem. 2007;282:10036–10046.
- Gonzalez-Lamothe R, El-Oirdi M, Brisson N, Bouarab K . The conjugated auxin indole-3-acetic acid-aspartic acid promotes plant disease development. Plant Cell. 2012;24: 762–777.
- Grubb CD, Abel S. Glucosinolate metabolism and its control. Trends Plant Sci. 2006;11:89–100.
- Denancé N, Ranocha P, Oria N, Barlet X, Rivière MP, Yadeta K, et al. Arabidopsis wat1 (walls are thin1)-mediated resistance to vascular pathogens is accompanied by cross-regulation of salicylic acid and tryptophan metabolism. Plant J. 2013;73:225–239.
- Vlot AC, Dempsey DM, Klessig DF. Salicylic acid, a multifaceted hormone to combat disease. Ann Rev Phytopathol. 2009;47:177–206.
- Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Ann Rev Phytopathol. 2005;43:205–227.
- Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, Oka N, et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature. 2012;486:228–232.
- Wu Y, Zhang D, Chu JY, Boyle P, Wang Y, Brindle ID, et al. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012;1:639–647.
- Choi SM, Song HR, Han SK, Han M, Kim CY, Park J, et al. HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid-mediated defense responses in Arabidopsis. Plant J. 2012;71:135–146.
- Wasilewska A, Vlad F, Sirichandra C, Redko Y, Jammes F, Valon C, et al. An update on abscisic acid signaling in plants and more. Mol Plant. 2008;1:198-217.
- Ton J, Flors V, Mauch-Mani B. The multifaceted role of ABA in disease resistance. Trends Plant Sci. 2009;14:310–317.
- Sánchez-Vallet A, Lopez G, Ramos B, Delgado-Cerezo M, Riviere MP, Llorente F, et al. Disruption of abscisic acid signalling constitutively activates Arabidopsis resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant Physiol. 2012;160:2109–2124.
- Inoue , Kinoshita T. Blue light regulation of stomatal opening and the plasma membrane H+ -ATPase. Plant Physiol. 2017;174:531–538.
- Eisenach C, de Angeli A. Ion transport at the vacuole during stomatal movements. Plant Physiol. 2017;174:520–530.
- Du M, Zhai Q, Deng L, Li S, Li H, Yan L, et al. Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell. 2014;26:3167–3184.
- Sierla M, Waszczak C, Vahisalu T, Kangasjärvi J. Reactive oxygen species in the regulation of stomatal movements. Plant Physiol. 2016;171:1569–1580.
- Clément M, Leonhardt N, Droillard MJ, Reiter I, Montillet JL, Genty B,et al. The cytosolic/nuclear HSC70 and HSP90 molecular chaperones are important for stomatal closure and modulate abscisic acid-dependent physiological responses in Arabidopsis. Plant Physiol. 2011;56:1481–1492.
- Noël LD, Cagna G, Stuttmann J, Wirthmüller L, Betsuyaku S, Witte CP, et al. Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. Plant Cell. 2007;19:4061–4076.
- Witte CP, Bhat R, Pochon N, Colby T, Parkerb JE. Interaction between SGT1 and Cytosolic/Nuclear HSC70 Chaperones Regulates Arabidopsis Immune Responses W. Plant Cell. 2007;19:401-406.
- Feng DX, Tasset C, Hanemian M, Barlet X, Hu J, Trémousaygue D, et al. Biological control of bacterial wilt in Arabidopsis thaliana involves abscisic acid signaling. New Phytol. 2012;194: 1035–1045.
- Jan AT, Azam M, Ali A, Haq Q. Novel approaches of beneficial Pseudomonas in mitigation of plant diseases an appraisal. J Plant Interact. 2011;6:195-205.
- Ebrahim, S, Usha K, Singh B. Pathogenesis Related (PR) Proteins in Plant Defense Mechanism. Sci Micro Pathogen. 2011;1043-1054.
- Van Loon LC, Van Strien EA. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol. 1999;55:85-97.
Citation: Belete T (2021) A Critical Review on Defense Mechanisms of Plants against Bacterial Pathogens: From Morphological to Molecular Levels. J Plant Pathol Microbiol. 12:534.
Copyright: © 2021 Belete T. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

